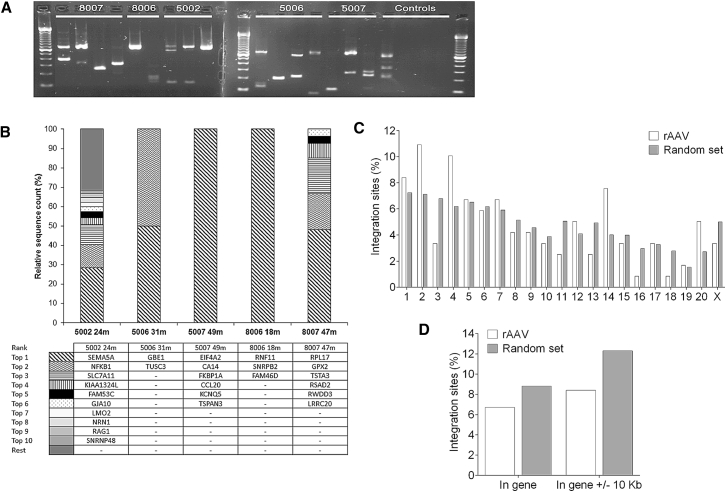
Figure 4.
Vector Integration Analyses
(A) Gel electrophoresis of LAM-PCR products retrieved from the different samples and negative controls (untransduced genomic DNA and three water controls) using MseI for restriction digest. Broken lines indicate two separate gels in the same image. Despite the presence of bands in the untransduced DNA control, due to homologies between the vector and genomic sequences, no amplification was obtained in the sequencing library preparation, indicating that only AAV-derived amplicons were used in later analyses. Due to AAV concatemers, which are also amplified by LAM-PCR, not all bands correspond to ISs. (B) Relative sequence counts of the ten most prominent ISs retrieved were calculated in relation to all uniquely mappable IS sequences. The RefSeq identity of the gene located adjacent to or at the IS are listed. (C) Chromosomal distribution of retrieved ISs was analyzed and compared to a synthetic random dataset of 8,628 ISs to determine eventual integration hotspots. (D) Distribution of the ISs within gene coding and nearby regions in comparison to a random dataset.