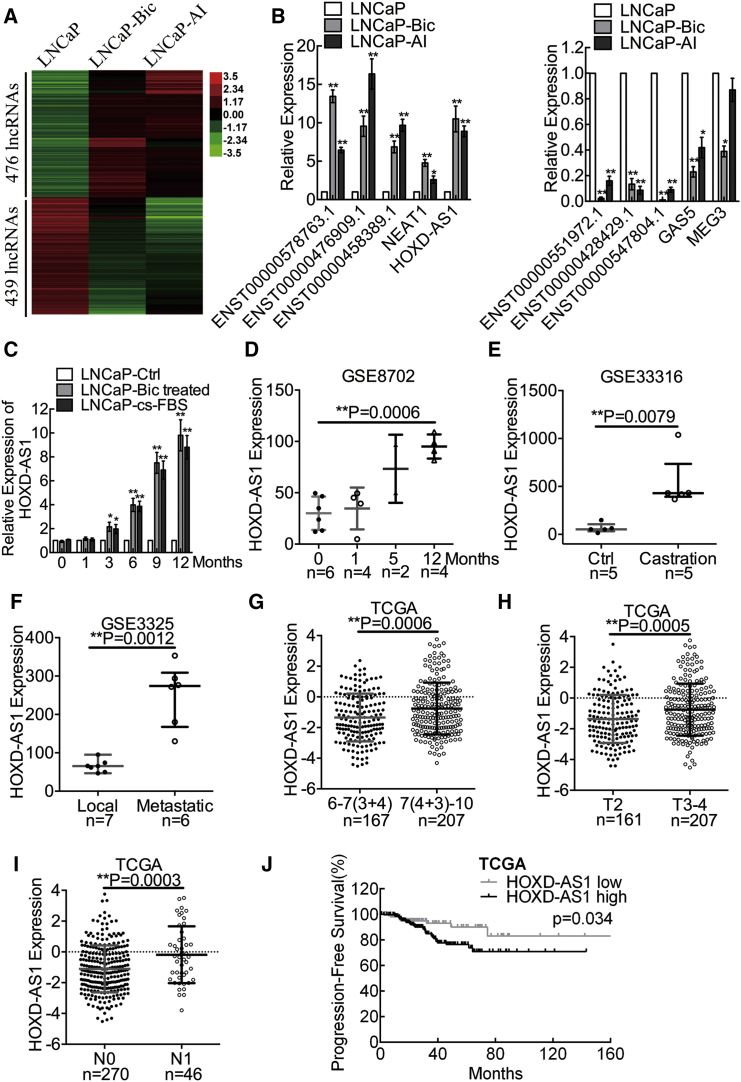
Figure 1.
HOXD-AS1 Is Identified as a Castration-Resistant Prostate-Cancer-Related lncRNA, Associates with Prostate Cancer Clinical Characteristics, and Predicts Disease Prognosis
(A) The differentially expressed lncRNAs in LNCaP versus LNCaP-Bic and LNCaP-AI were detected using a microarray. (B) The results from microarray analysis were validated by real-time qPCR. The results are presented as the means ± SD of values obtained in three independent experiments. (C) The expression of HOXD-AS1 in LNCaP cells treated with either bicalutamide or androgen ablation at different points in time was detected by real-time qPCR. The results are presented as the means ± SD of values obtained in three independent experiments. (D) GEO analysis of HOXD-AS1 expression in LNCaP cells under androgen ablation. The whiskers indicate means ± SD in the plots. (E) GEO analysis of HOXD-AS1 expression in castrated mice xenografts. The whiskers indicate median ± quartile in the plots. (F) GEO analysis of HOXD-AS1 expression in metastatic PCa versus localized PCa. The whiskers indicate medians ± quartile in the plots. (G) The expression of HOXD-AS1 in Gleason score 6–7(3+4) versus Gleason score 7(4+3)–10 PCa from TCGA database. The whiskers indicate means ± SD in the plots. (H) The expression of HOXD-AS1 in T2 versus T3-4 PCa. The whiskers indicate means ± SD in the plots. (I) The expression of HOXD-AS1 in N0 versus N1 PCa. The whiskers indicate means ± SD in the plots. (J) The progression-free survival rates of the 309 PCa patients were compared in the HOXD-AS1-low and HOXD-AS1-high groups. −1.8 was used as cutoff value in the survival analysis. The total number of patients was 374, 368, and 316 in each TCGA analysis, respectively. Patients with unavailable profiles were excluded before each analysis. See also Figure S2. *p < 0.05; **p < 0.01.