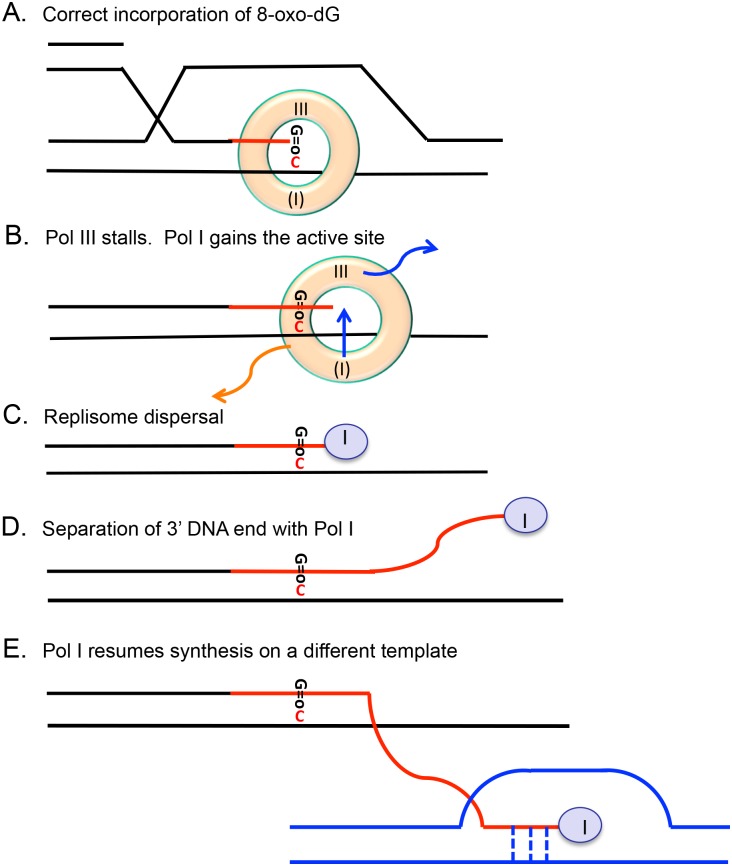
Fig 8. A model for template switching leading to chromosomal rearrangement following incorporation opposite 8-oxo-dG.
(A) Pausing of the replication complex after Pol III places 8-oxo-dG opposite C [38] (shown, or after Pol III stops after placing C opposite to 8-oxo-G in the template strand, not illustrated) sometimes leads to (B) exit of Pol III from active position in the β-clamp, and the DNA polymerase from the inactive position (Pol I here) is switched into the active site. (C) Prolonged pausing of the replisome leads to dispersal of the β-clamp. (D) The Pol I-bound primer end can become detached, possibly by activation of the editing function, so that template switching can occur, initiating GCR by MMBIR [45, 46]. (E) In this case the primer binds to a template at a D-loop at a non-homologous position at microhomology (green) or any other single-stranded DNA (not illustrated). Extension by Pol I will stabilize the junction allowing non-homologous recombination, which creates gene duplications, deletions and other GCRs. Conventions are as in Fig 7 except that the orange arrow indicates detachment of the β-clamp, and the vertical dashed bars in part (E) indicate a microhomologous junction.