Figure 2.
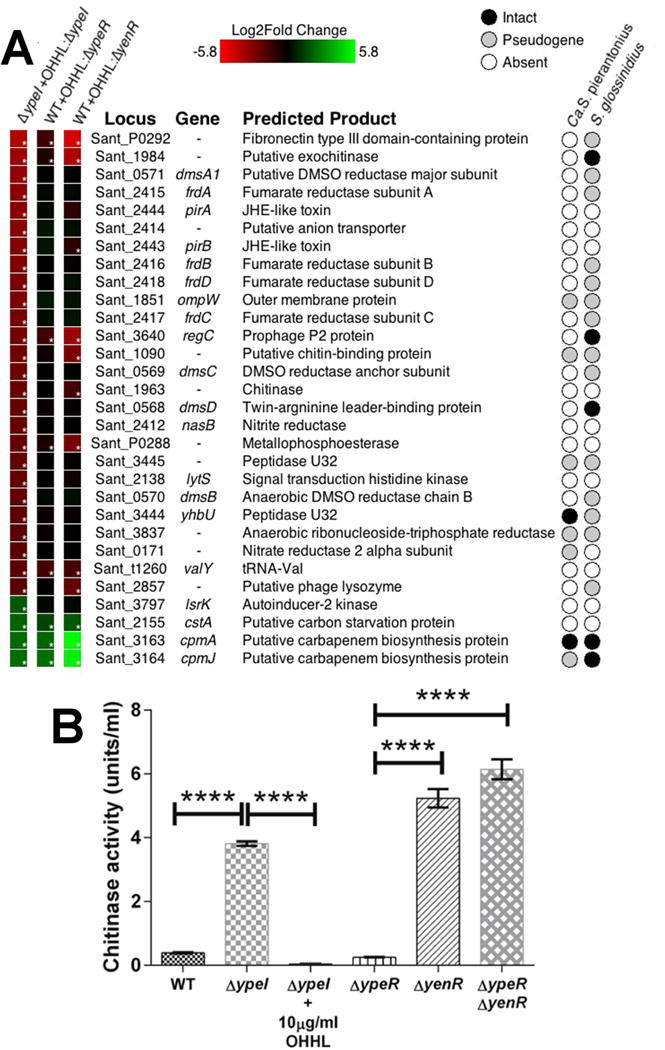
Transcriptomic (RNA-Seq) Analysis of QS-regulated genes in S. praecaptivus. (A) Heat maps showing changes in gene expression. The first column shows changes in the 30 most differentially expressed genes in the S. praecaptivus ΔypeI strain in response to addition of exogenous OHHL. The second and third columns show changes in the expression of the same 30 genes in a WT strain (supplemented with OHHL) relative to ΔypeR and ΔyenR strains (respectively), lacking exogenous OHHL. The asterisks in the boxes on the left indicate statistically significant levels of differential expression. The matrix on the right hand side of the panel reveals the status of the 30 genes in Sodalis-allied insect symbionts that are closely related to S. praecaptivus. Complete transcriptomic datasets are provided in Table S1. (B) Chitinase activities of WT and mutant S. praecaptivus strains, in the presence or absence of OHHL. All samples were assayed in triplicate and error bars show standard deviations. One way ANOVA comparisons between genotypes: ΔypeI vs. ΔypeI supplemented with 10 μg/ml OHHL, WT vs. ΔypeI, ΔypeR vs. ΔyenR and ΔypeR vs. ΔypeRyenR, all yield p <0.0001; ΔyenR vs. ΔypeR ΔyenR double mutant yields p = 0.0006.
