Figure 3.
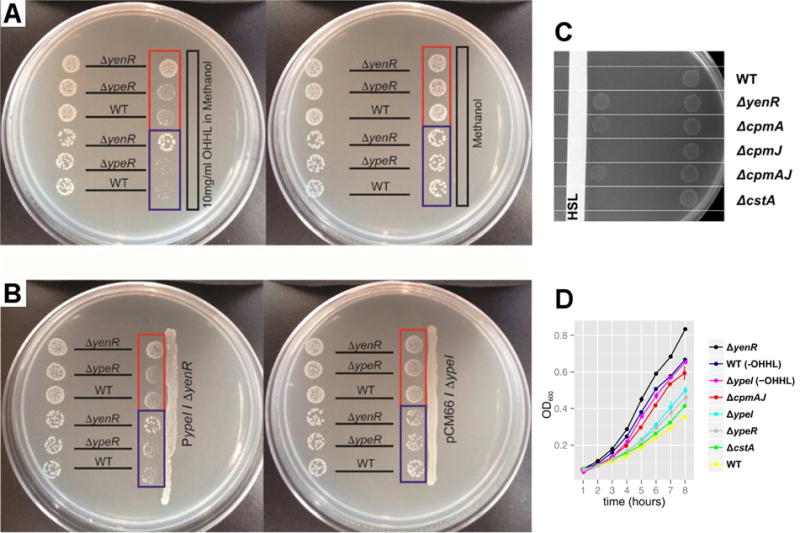
QS Induces Growth Suppression in S. praecaptivus. (A & B) Each bacterial strain (labeled according to genotype) was spotted in two positions on the plate. Panel A shows spots placed either distal (left) or proximal (right) to (A) a strip of sterile paper that was impregnated with exogenous OHHL in methanol (left plate) or methanol alone (right plate). Panel B shows spots placed either distal (left) or proximal (right) to a streak of the S. praecaptivus ΔyenR strain maintaining plasmid pCM66 overexpressing the ypeI gene (left plate) or a streak of the S. praecaptivus ΔypeI strain maintaining plasmid pCM66 lacking ypeI (right plate). The spots highlighted in the red boxes have a 10-fold higher concentration of cells than their counterparts highlighted in blue. (C) S. praecaptivus strains (labeled according to genotype) were spotted in two positions on a plate, either proximal (left) or distal (right) to a strip of sterile paper impregnated with exogenous OHHL in methanol (labeled “HSL”). Note that deletion of cpmA alone relieves QS-mediated growth suppression. Deletion of either cpmJ or cstA (another gene whose transcription is increased under quorum), has no effect on growth rate. (D) Assays of growth performed in liquid media containing 1 mg/ml OHHL, or no HSL as indicated by the “-OHHL” suffix. Data were obtained from three biological replicates and error bars show standard errors. Note that growth is enhanced by deletion of yenR or cpmAJ.
