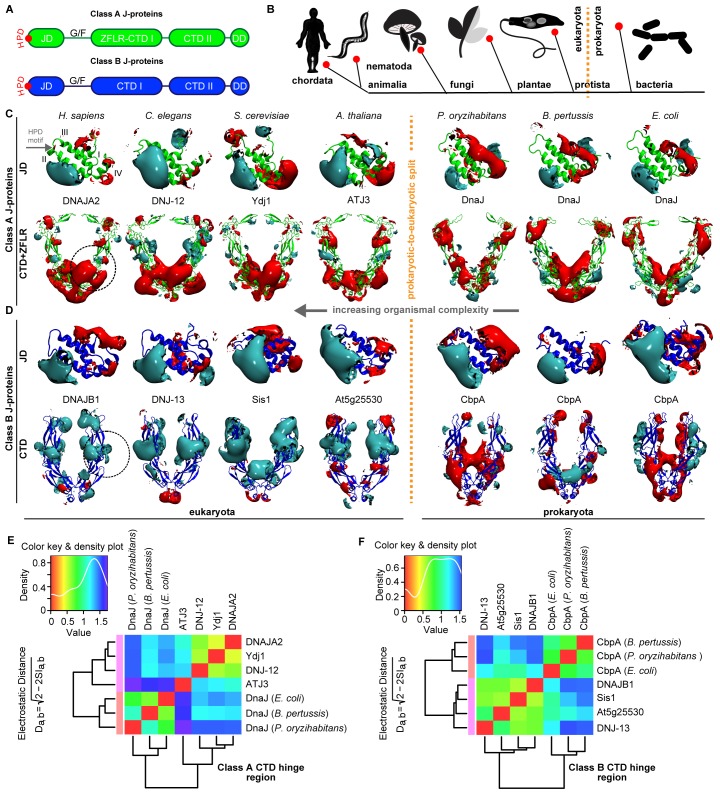
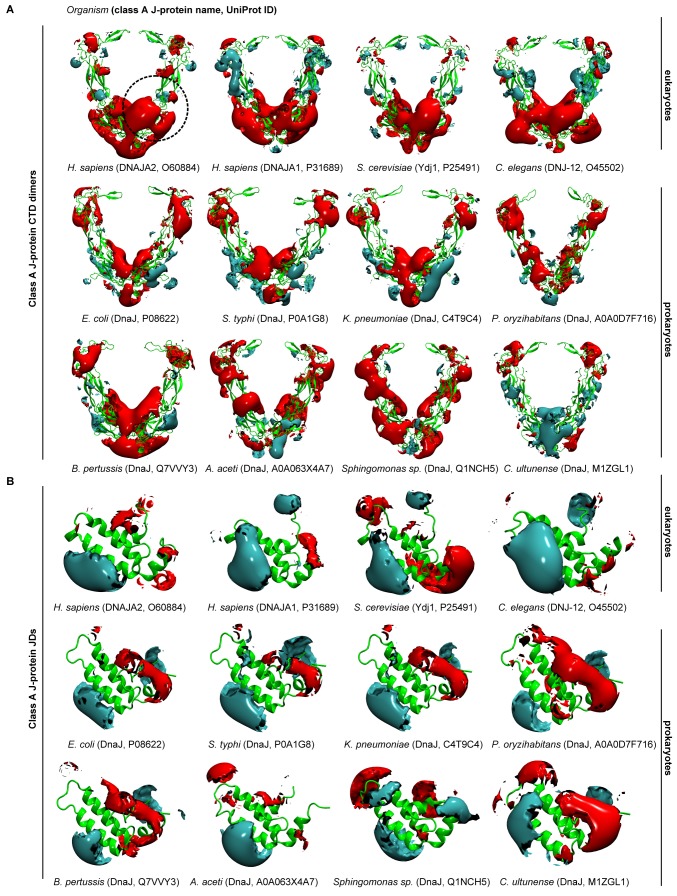
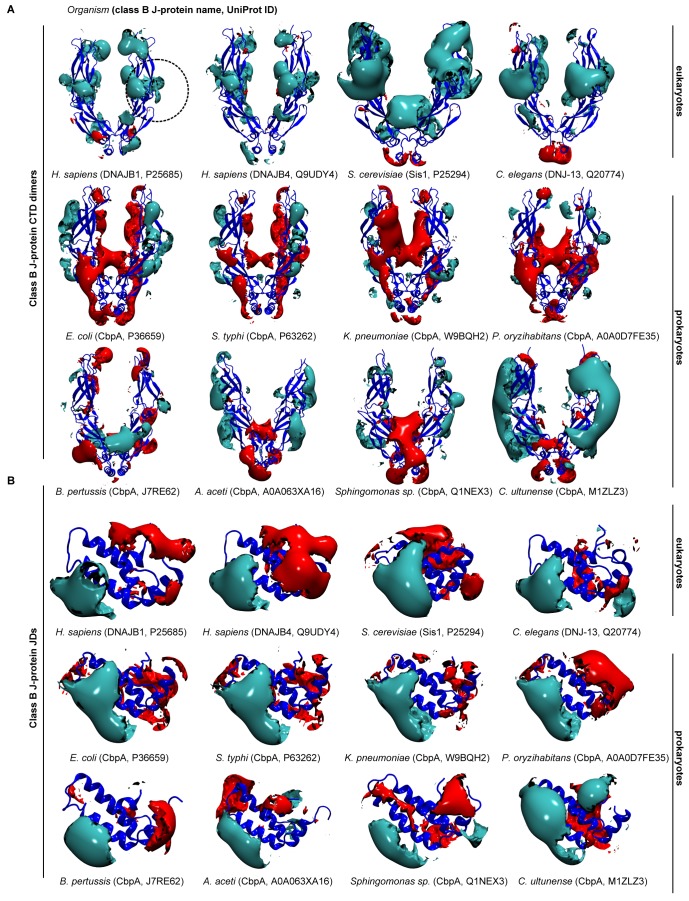
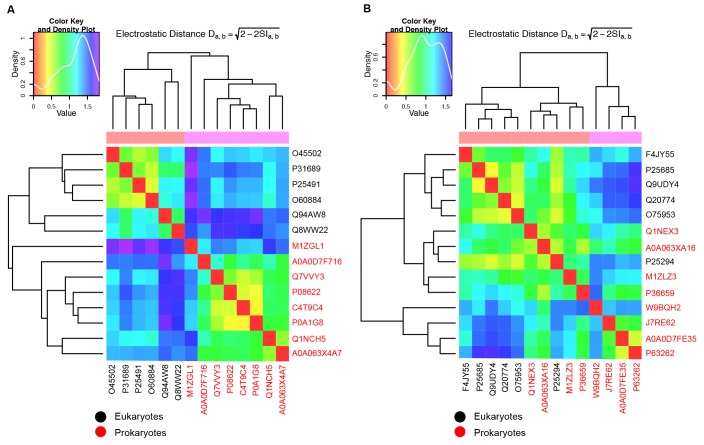
Figure 1. Conservation of class-specific electrostatic potential distributions predict interclass J-protein complex formation in eukaryotes.
(A) Domain architecture of class A and class B J-proteins (shown as protomers). Class A J-proteins have an N-terminal J-domain (JD), a glycine/phenylalanine-rich flexible region (G/F), C-terminal β-sandwich domains (CTD-I and II) and a CTD-I inserted zinc-finger-like region (ZFLR). The Hsp70-interacting HPD motif is indicated in red. Protomer dimerization to form homodimers occurs at the dimerization domain (DD). The ZFLR in CTD-I is absent in the domain architecture of class B J-proteins. (B) Evolutionary tree ranking the kingdoms of organisms analyzed in this study. (C, D) Electrostatic isopotential contour maps (cyan + 1, red −1 kcal/mol/e) of CTD homodimers and of J-domains of class A (green cartoon diagrams) (C) and class B (blue cartoon diagrams) (D) J-proteins. Roman numerals show the four α-helices on class A JD. J-proteins from the following organisms are compared: Homo sapiens (DNAJA2, DNAJB1), Caenorhabditis elegans (DNJ-12, DNJ-13), Saccharomyces cerevisiae (Ydj1, Sis1), Arabidopsis thaliana (ATJ3, At5g25530), Pseudomonas oryzihabitans, Bordetella pertussis and Escherichia coli (DnaJ, CbpA). The dashed circles on the CTDs of DNAJA2 and DNAJB1 represent the spherical region used for local PIPSA analysis of electrostatic potential similarity. (E, F) Local PIPSA analysis results for class A CTD (E) and class B CTD (F) electrostatic potentials. The electrostatic potentials in the spherical regions (radius of 25 Å) indicated by the dashed black circles in (C) and (D) were clustered by similarity using Ward’s clustering. The heat maps show clustering of J-proteins by similarity (higher similarity indicated by a red shift).