Figure 2. Phylogenetic and coevolutionary analyses of the JD-CTD interaction between class A and B J-proteins.
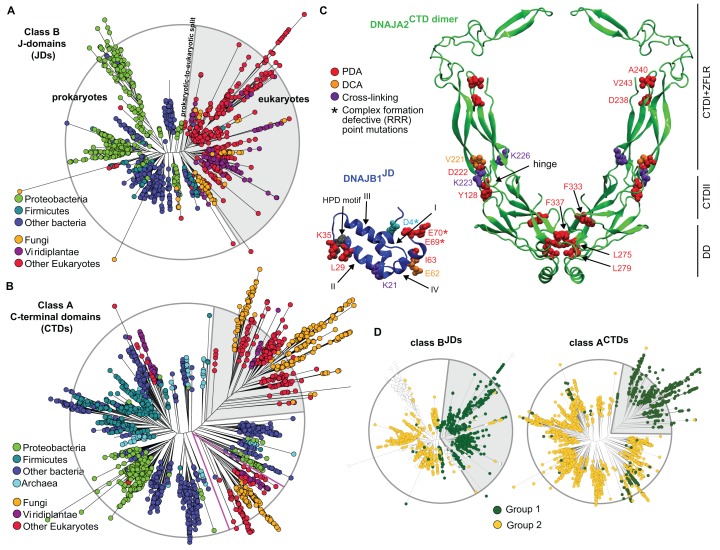
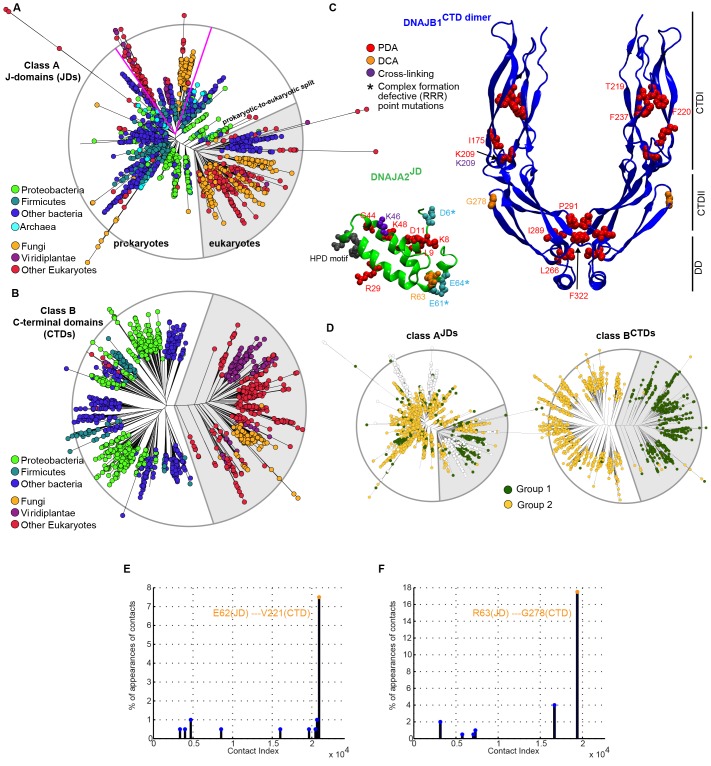
(A) Phylogenetic tree of class B J-domains. Color-coding separates different phylogenetic groups. Grey area highlights the separation of eukaryotes (fungi, viridiplantae and other eukaryotes) from prokaryotes (proteobacteria, firmicutes and other bacteria) and Archaea. (B) As in (A), phylogenetic tree of class A CTDs. Pink lines delimit organellar sequences of eukaryotic organisms. (C) Structural view of most discriminating positions predicted by PDA (red) plotted on JD of DNAJB1 (blue, five positions) and CTD of DNAJA2 (green, nine positions). DCA-derived coevolving residue pairs depicted on DNAJB1JD and DNAJA2CTD (orange). Experimentally determined cross-linking residues between DNAJB1 and the DNAJA2 are indicated in purple (Nillegoda et al., 2015). Location of the triple charge reversion (E/D→R) mutations that disrupts interclass J-protein complex formation between DNAJB1 and DNAJA2 denoted by (*) (Nillegoda et al., 2015). The HPD motif of DNAJB1JD is shown in grey. Roman numerals show the four α-helices on JD. (D) Mapping of sequence clustering derived from PDA (see Materials and methods) using the most discriminating positions on to JD and CTD trees of class B and class A J-proteins, respectively. The two identified groups (green and yellow nodes) covered 81% in the case of the clustering done on the JDs and 100% when clustering was done on the CTDs. Unclassified sequences are depicted in white.