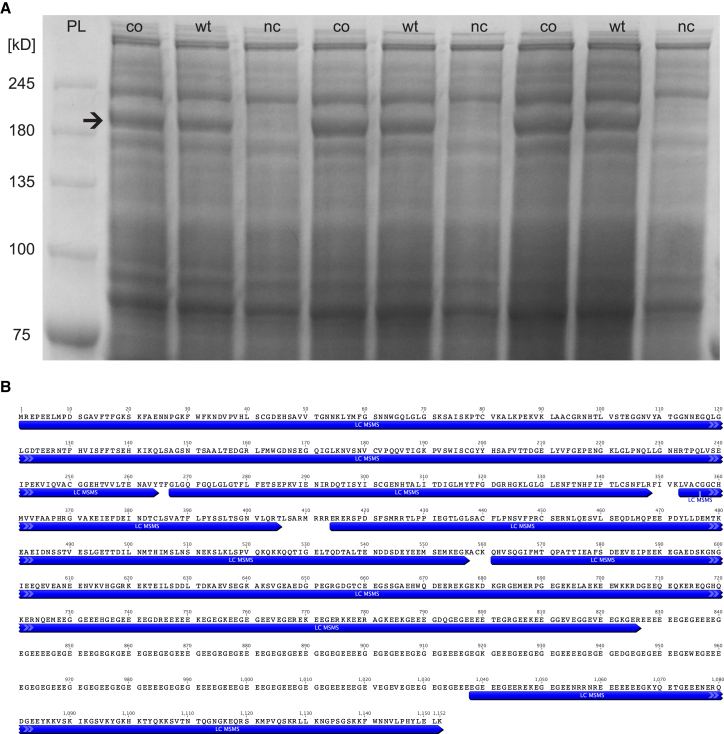
Figure 2.
Liquid Chromatography-Tandem Mass Spectrometry of wtRPGRORF15 and coRPGRORF15
(A) Following overexpression in HEK293T cells, wtRPGRORF15 and coRPGRORF15 were purified from whole cell lysate by SDS-PAGE. In each lane, a single band above 180 kDa was identified as RPGRORF15 (arrow). This band was missing in the negative control (nc) lanes. No additional bands are visible, which would be different between WT and coRPGRORF15 lanes. (B) Identified RPGRORF15 bands were then excised for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. More than 80% of peptide sequence could be directly identified (indicated by dark arrows below amino acid sequence). Unidentified amino acids (no arrows) are largely based in the ORF15 sequence, which escapes LC-MS/MS analysis because of its repetitive, glutamic acid and glycine-rich sequence. Note that the C-terminal sequence following the ORF15 region was identified, which rules out premature termination of translation. Together with the identical migration pattern between the wtRPGRORF15 and coRPGRORF15 samples in the SDS-PAGE (A), this suggests that codon optimization does not result in alternative splicing, but in a protein product equivalent to the wtRPGRORF15.