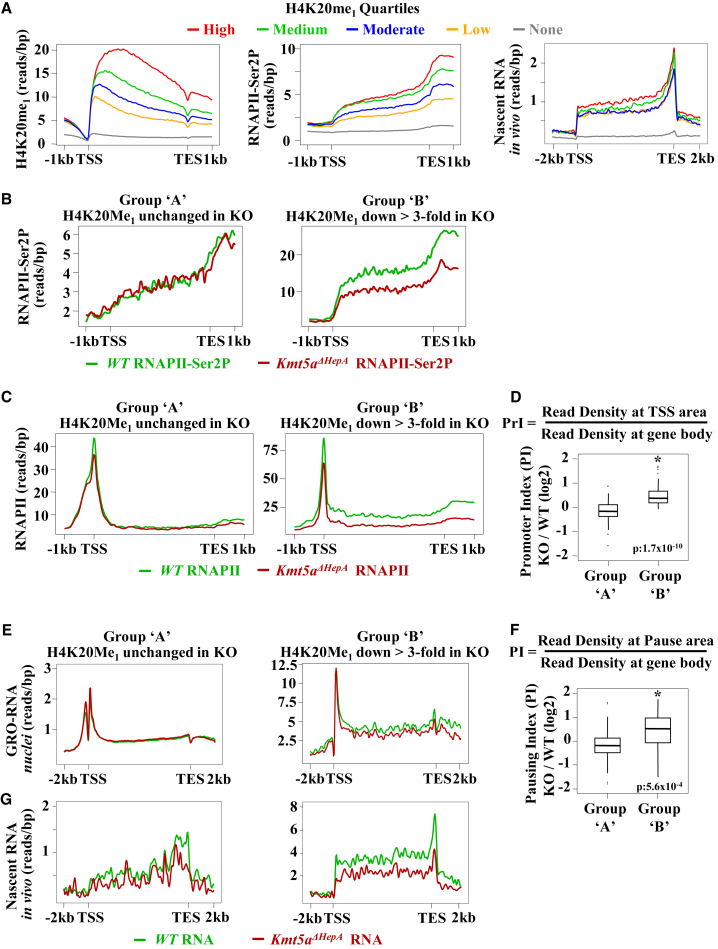
Figure 2.
H4K20 Monomethylation Correlates with RNA Pol II Distribution on Active Genes
(A) Average H4K20Me1 (left panel), RNA Pol II-Ser2 (middle panel), and in vivo ethynyl-uridine (EU)-labeled nascent RNA (right panel) coverage profiles at the genes expressed in P45 wild-type mouse livers. Genes containing H4K20Me1 in their gene bodies (n = 6,570 genes with normalized reads per gene length > 0.03 bp) were divided into quartiles (high, red line, n = 1,643; medium, green line, n = 1,642; moderate, blue line, n = 1,642; and low, orange line, n = 1,643) according to H4K20Me1 read densities. The distribution over non-methylated, annotated genes (n = 11,472 genes) is shown by the gray line. The graphs show average coverage of ChIP-seq reads obtained with H4K20Me1 and RNA Pol II-Ser2 antibodies and EU-labeled nascent RNA reads over the gene body regions.
(B and C) Distribution of RNA Pol II-Ser2 (B) and total RNA Pol II (C) in the group of genes displaying low (group A) and high (group B) H4K20Me1 methylation turnover. The graphs show average coverage of ChIP-seq reads.
(D) Quantitative comparison of changes in PrIs in hepatic genes displaying different methylation turnover. The PrI was calculated as the ratio of the normalized RNA Pol II coverage (reads/bp) of a 500 bp area around the TSS of mouse genes divided by the window length (500 bp) to the normalized RNA Pol II coverage over the rest of the gene body. Boxplots depict changes in PrI values between wild-type and Kmt5aΔHepA mice (knockout [KO]) in group A and group B genes as indicated. Statistical significance was assessed by Welch’s t test.
(E) Distribution of nascent RNA reads labeled by Bromo-Uridine (BrU) in isolated nuclei evaluated by GRO-seq (GRO-RNA) in group A and group B genes. Note the different scales of the y axis.
(F) Pausing index (PI) calculated as the ratio of the normalized nascent RNA coverage (reads/bp) in the TSS to the +50 nt area of mouse genes divided by the window length (500 bp) to the normalized nascent RNA coverage over the rest of the gene body. The results are presented in boxplots as in (D).
(G) Distribution of in vivo nascent RNA reads by EU (nascent RNA in vivo) in group A and group B genes. Note the different scales of the y axis.
See also Figure S2.