Fig. 5.
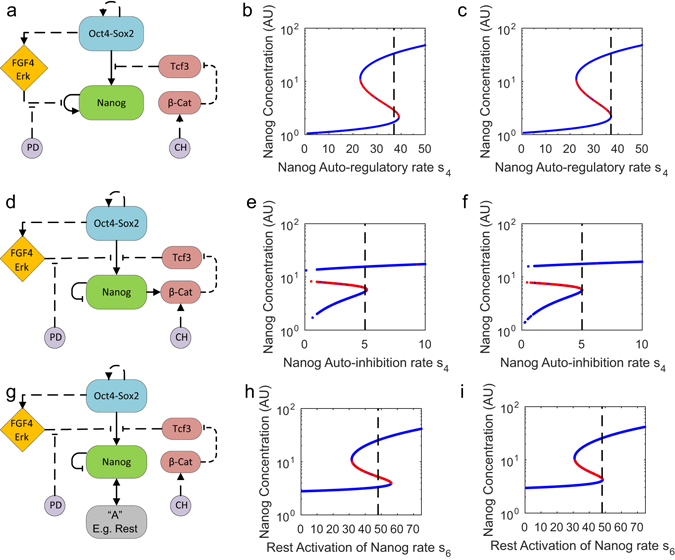
Simpler network models demonstrating bistability of Nanog. a Scheme of simplified network, composed of core factors Nanog, Oct4, Sox2; Fgf4/Erk, β-catenin and Tcf3 are added to account for the inputs (i.e., PD and Chiron (CH), the two inhibitors present in 2i/LIF). Solid and dashed lines indicate transcriptional and non-transcriptional interactions, respectively. b, c Continuation of Nanog steady-state for the deterministic system (no noise) in serum/LIF (b) and 2i/LIF (c). Blue lines are stable steady states, while red lines are unstable. The maximal transcription rate, s 4 (dotted black line), intersects both NH and NL steady-states, indicating bistability in serum/LIF (b), but is close to the saddle-node bifurcation in 2i/LIF (c). d Scheme of Nanog auto-inhibitory simplified network, with additional activation of β-catenin by Nanog. e, f Continuation of Nanog steady-state for the deterministic system (no noise) in serum/LIF (e) and 2i/LIF (f). Blue lines are stable steady-states, while red lines are unstable. The maximal transcription rate of Nanog auto-inhibition, s 4 (dotted black line), intersects both NH and NL steady-states, indicating bistability in serum/LIF (e), while being closer to the saddle-node bifurcation in 2i/LIF (f). g Scheme of Nanog auto-inhibitory simplified network, with additional positive feedback supplied via an extra factor, i.e. Rest. h, i Continuation of Nanog steady-state for the deterministic system (no noise) in serum/LIF (h) and 2i/LIF (i). Blue lines are stable steady-states, while red lines are unstable. The maximal transcription rate of Rest activation on Nanog, s6 (dotted black line), intersects both NH and NL steady-states, indicating bistability, in serum/LIF (h), and is very close to the saddle-node bifurcation in 2i/LIF (i)
