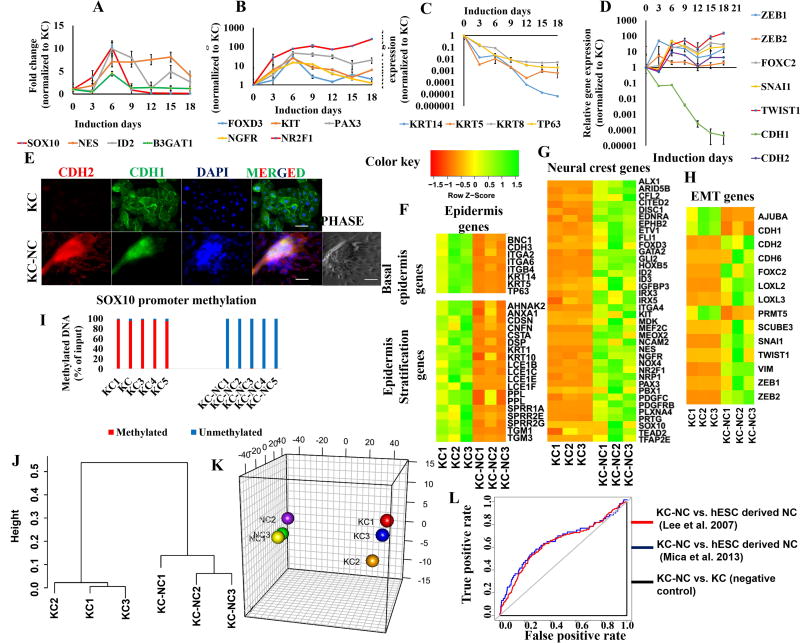
Figure 2. KC-NC display global molecular characteristics of NC cells.
KC-NC express NC specific genes (SOX10, NES, PAX3, FOXD3, NGFR, KIT, ID2, B3GAT1, NR2F1) transiently during induction (n=3 donors) (A, B) and downregulate KC specific genes (KRT14, KRT5, KRT8, and TP63) (n=3-5 donors) (C). KC-NC induction is marked by upregulation of EMT genes (ZEB1, ZEB2, SNAI1, FOXC2 and TWIST1) including classical cadherin switch (CDH1 to CDH2) (n=3-5 donors) (DE). Transcriptome sequencing (RNA-Seq) analysis of KC-NC and their respective parental KC (n=3 donors) shows basal epidermis and epidermal stratification genes were downregulated in KC-NC (F) while NC specific genes (G) and EMT genes (H) were upregulated. The genomic region known to regulate SOX10 transcription was severely hypomethylated in KC-NC, while heavily methylated in KC (methylation: 97±1.2% of input genomic DNA for KC and 0.54±0.3% of input genomic DNA for KC-NC, p = 5.45x10−15, n=5 donors) (I). Hierarchical clustering performed on all pairwise distances between the global transcriptome-wide RNA-seq profiles of KC and respective KC-NC shows distinct clustering between the two groups (J). Three-dimensional multi-dimensional scaling (3D-MDS) of all differentially expressed genes demonstrates that KC and KC-NC populations clustered separately (K). KC-NC displayed molecular signatures similar to human embryonic stem cell derived NC cells as confirmed by AUC (area under curve) of ROC plots (L). Statistical significance of ROC tests was confirmed using Wilcoxon rank-sum test. ([16], AUC: 0.597; p=4.304x10−5[1] AUC: 0.644; p= 5.454 x10−5 and parental KC AUC: 0.0003461405; p= 1). Scale bars, 50µm. All values are mean±SD. Experiments were repeated three times independently.