Fig. 2.
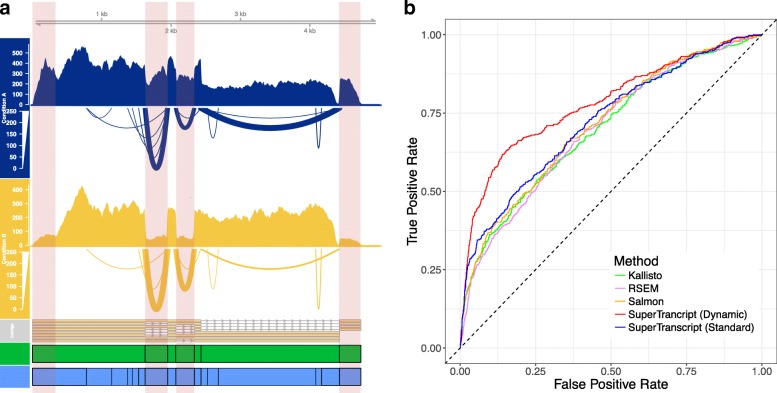
a An example of gene visualisation for a de novo assembled transcriptome. We show the read coverage and splice junctions of Condition A (blue) and Condition B (yellow), the controls and knockdowns, respectively. Differential isoform usage between two samples of different conditions can be seen in the read coverage (highlighted regions). The two alternative block annotations for the superTranscript – Standard (green) and Dynamic (blue) – are illustrated underneath. Dynamic block boundaries are located at splice junctions (defined by at least five spliced reads). b Receiver operating characteristic curve for detecting differential isoform usage using de novo assembled transcripts from the Trapnell et al. dataset. True and false positives are defined using a reference genome analysis (See ‘Methods’)