Figure 3.
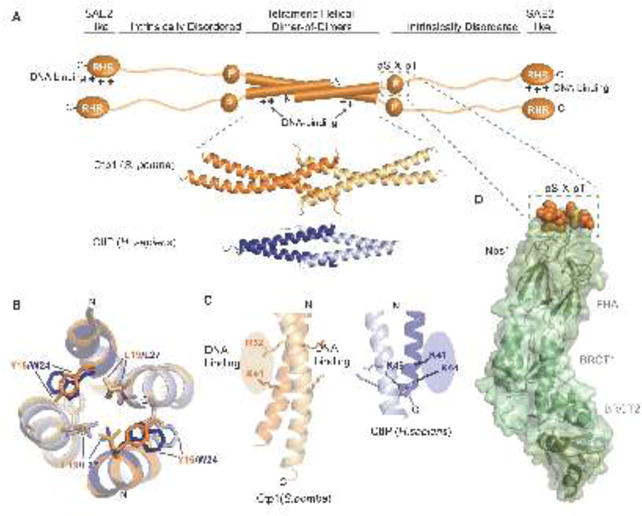
Structural features of Ctp1 and CtIP. (A) Domain organization of Ctp1 (S. pombe) and CtIP (H.sapiens). The domains of Ctp1 (orange schematic) highlight the core N-terminal tetrameric helical dimer of dimers (THDD), an intrinsically disordered region that contains both the phosphorylated pS-x-pT motif for Nbs1-binding (green dotted box) and the C-terminal SAE2-like domain. Crystal structures of Ctp1 (orange; PDB 4X01) and CtIP (blue; PDB 4D2H) show the conserved tetramerization core formed by interlocking alpha-helices. (B) Comparison of Ctp1 and CtIP tetramerization. Overlay of Ctp1 (orange/yellow) and CtIP (blue/gray) tetramerization interface, mediated by leucine and aromatic amino acids. (C) DNA binding surfaces. Ctp1-DNA interactions map to the exposed surface of Ctp1 dimers (orange). A similar basic surface in CtIP also exists. (D) Ctp1-Nbs1 complex. The phosphorylated Ctp1 (orange) binds a positively charged, surface-exposed phosphoprotein recognition pocket of the Nbs1 FHA domain (green) (PDB 3HUF).
