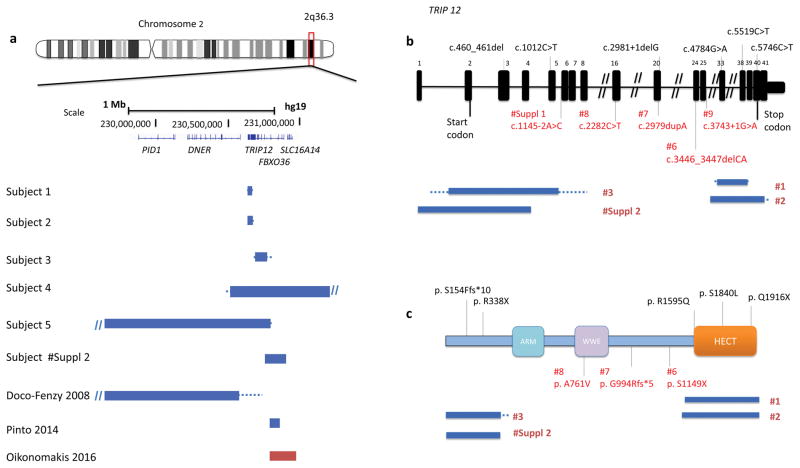
Figure 1.
(a) Schematic representation of 2q36.3 CNV deletions involving TRIP12 identified by exon-targeted CMA in six subjects from this study and three CNV subjects reported previously. Deletions are indicated in blue and duplications in red. The dotted lines depict the maximum deletion regions. (b) TRIP12 gene exonic structure. Three TRIP12 exonic deletion CNVs identified in subjects 1–3 are indicated under the diagram (blue bars indicate the minimum deletion region and the dotted lines depict the maximum deletion regions). The variants from this study are indicated in red and the previously reported variants are indicated in black. Reported variants include two missense c.5519C>T (p.S1840L) and c.4784G>A (p.R1595Q) and two truncating c.2981+1delG and c.1012C>T (p.R338X) changes from ASDs cohort studies (O’Roak et al. 2014) and c.5746C>T (p.Q1916X) and c.460_461del (p.S154Ffs*10) from 2,104 trio studies (Lelieveld et al. 2016). (c) Functional domains of TRIP12 include ARM domain (turquoise), WWE domain (purple) and HECT (E6AP-type E3 ubiquitin-protein ligase) (orange).