Figure 6.
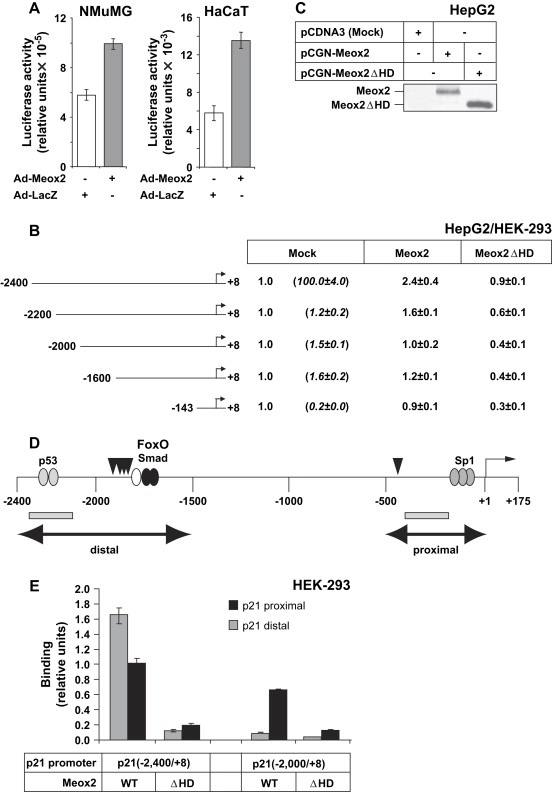
Meox2 induces the p21 promoter–enhancer via its distal region. (A) Transactivation of the −2400/+8p21 promoter in NMuMG and HaCaT cells transiently infected with Ad‐Meox2 or Ad‐LacZ (MOI 100) for 24h. Normalized relative luciferase activity is shown in arbitrary units. (B) Luciferase p21 promoter assays of the indicated promoter deletion constructs in HepG2 and HEK‐293 cells transfected with mock vector (pcDNA3) or specific Meox2‐expressing vectors (wild type, Meox2, and homeodomain deletion mutant, Meox2ΔHD). Values express average normalized relative luciferase activity and standard deviations from triplicate determinations. Similar data were obtained from both cell lines. All basal promoter activities (Mock) are shown as 1, relative to which the promoter activities obtained after Meox2 transfections are calculated. The differences in basal promoter activity between the various deletion constructs are shown in parenthesis under Mock. (C) Immunoblot analysis of transfected wild type Meox2 and mutant lacking its homeodomain (ΔHD) in cell lysates used for the luciferase assay of panel B, using the anti‐HA antibody. (D) Diagram of the human p21 promoter–enhancer indicating the distal and proximal regions (double‐headed arrows) with the corresponding p53, FoxO3a‐Smad and Sp1 binding elements, and the transcriptional initiation (+1) site. Vertical arrowheads indicate the putative Meox2‐binding sites and the PCR fragments amplified during ChIP analysis are shown as grey rectangles. (E) ChIP analysis of the human p21 promoter–enhancer in transfected HEK‐293 cells with the two indicated p21 genomic fragments and wild type (WT) Meox2 expression vector. The homeodomain deletion mutant Meox2(ΔHD) serves as negative control. Q‐PCR assays measure genomic sequences corresponding to a distal and a proximal p21 genomic fragment as illustrated in panel D (grey rectangles).
