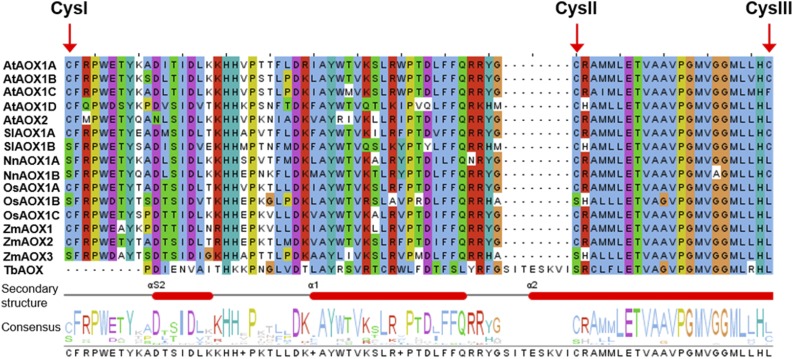
Figure 1.
Section of a multiple sequence alignment of AOX isoforms. Amino acid sequences of AOX isoforms from different plant species were aligned using the bioinformatic tools Clustal Omega and Jalview (Waterhouse et al., 2009; Sievers et al., 2011). The region including all Cys residues of plant AOX isoproteins is shown (for total alignment, see Supplemental Fig. S1). The top five sequences show AOX isoforms from Arabidopsis (AtAOX). The following lines illustrate sequences of tomato (SlAOX), lotus (NnAOX), rice (OsAOX), and maize (ZmAOX). The last line represents AOX from Trypanosoma brucei (TbAOX). Note that SlAOX1B, NnAOX1A, NnAOX1B, OsAOX1B, and ZmAOX3 possess a Ser residue at the position of CysI of AOX isoforms from Arabidopsis.