Figure 3. Illustrations of yeast genomic methods for target-identification and mechanism-of-action studies.
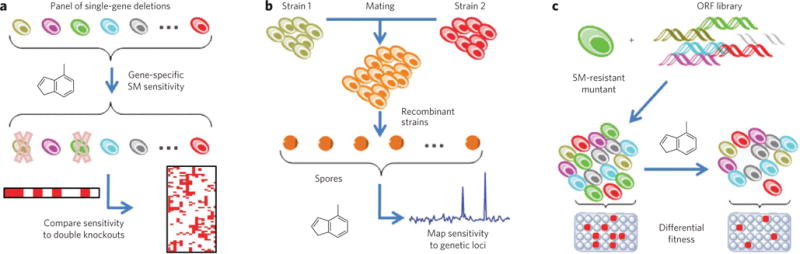
(a) A panel of viable single-gene deletions is tested for small-molecule (SM) sensitivity, indicating synthetic-lethal interactions between potential targets and the original deletion; mechanisms are interpreted by comparing interactions to double-knockout strains146. (b) Different strains of diploid yeast are mated to form F1 recombinants, and meiotic progeny are subjected to small molecules; segregation frequencies allow mapping of small-molecule sensitivity to genetic loci147. (c) A recessive small molecule–resistant mutant is transformed with a wild-type open reading frame library; transformants obtaining a wild-type copy of the mutant gene are selectively sensitive to small molecules, resulting in their depletion among pooled transformants, as quantified by microarray89.
