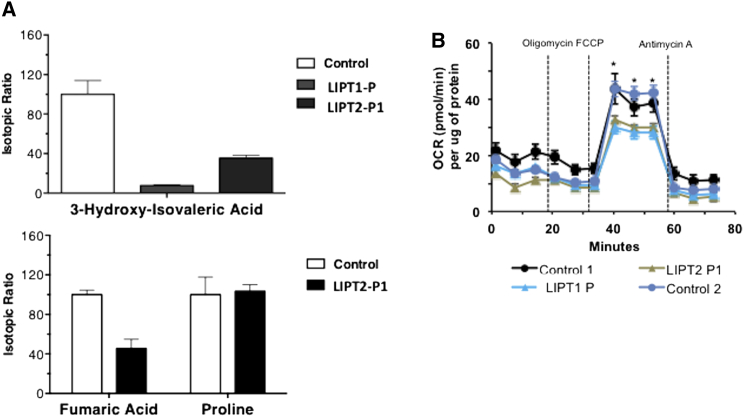
Figure 2.
Labeled Glutamine and Leucine Loading Tests and Oxygen Consumption Rates in Fibroblasts from Control Subjects and Individuals with LIPT1 and LIPT2 Deficiencies
(A) Labeled to natural ratios for 3-hydroxyisovaleric acid after a 13C6 leucine loading test in fibroblasts of an individual with LIPT1 deficiency and LIPT2-P1 and for fumaric acid and proline after a 13C5 glutamine loading test in LIPT2-P1 and control fibroblasts. Labeled leucine and glutamine loading tests are consistent with decreased BCKDHc activity and Krebs cycle activity defects. Labeled amino acids were acquired from Eurisotop. Organic acids derived from labeled substrates were measured by gas chromatography-mass spectrometry (GC-436 Scion-TQD, Brüker Daltonics). Results are presented as means ± SD of triplicates.
(B) Oxygen consumption rates measured in fibroblasts from healthy subjects and individuals with LIPT1 and LIPT2 deficiencies. Basal OCR levels did not differ significantly in fibroblasts from healthy control subjects and individuals with LIPT17 and LIPT2 deficiencies. By contrast, when challenged with a mitochondrial uncoupler (FCCP), fibroblasts from healthy individuals responded with the expected increase in oxygen consumption, whereas the response in cells from individuals with LIPT1 and LIPT2 deficiency was significantly lower. OCR was measured using the XF Cell Mito Stress Test Kit and XFe96 analyzer (Seahorse Bioscience, Agilent Technologies) following the manufacturer’s protocols.23 Cells were seeded at the density 30,000 cells/well in an XFe96cell culture microplate and allowed to attach for 3 hr before the measurement. Basal OCR was measured, followed by sequential treatment with Oligomycin A (1 μM), FCCP (1 μM), and Antimycin A (1 μM). Each treatment was measured every 3 min (3 min measurement) three times and a minimum of six replicates were utilized per condition. All compounds and materials were obtained from Seahorse Bioscience. Protein concentrations in each well were determined with the BCA method (Pierce) in cell lysates after the measurement. Data are presented as mean ± SEM normalized to protein content in each well. Statistical test was performed using ANOVA test; ∗p ≤ 0.05 control versus affected individuals’ cells.