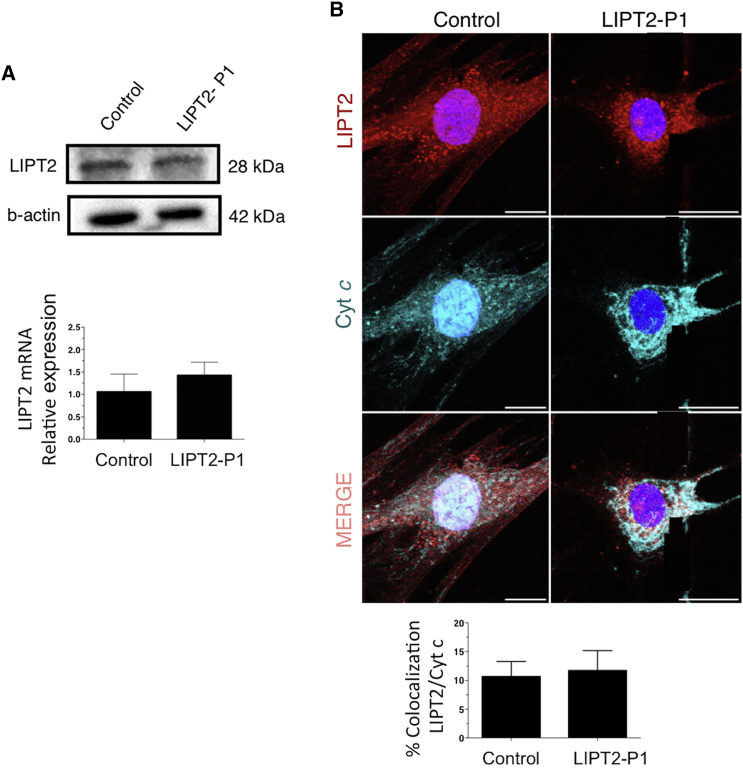
Figure 4.
Immunoblot Analysis, mRNA Expression, and Immunofluorescence of LIPT2 in Fibroblasts
(A) Fibroblasts of LIPT2-P1 and control contain similar amount of LIPT2 protein and mRNA. LIPT2 protein level was studied three times (anti-LIPT2 antibody: ab173981 Abcam, 1:500; anti-β-actin: Sigma-Aldrich cat# A5441, RRID: AB_476744, 1:10,000). LIPT2 mRNA expression was analyzed by quantitative PCR and normalized to the β-actin mRNA level (ABI PRISM 7300 Sequence Detection System instrument; TaqMan Universal PCR Master Mix from Applied Biosystem). Data are presented as mean ± SD of triplicates. For studies in LIPT2-P1 fibroblasts, primer sequences are: forward primer LIPT2: 5′-CGT GGT TTG AGC ACA TCG-3′; reverse primer LIPT2: 5′-AAG GCC ACA AGG AAA GGT G-3′.
(B) Fibroblasts from LIPT2-P1 and control exhibited colocalization of cytochrome c and LIPT2. Rabbit anti-LIPT2 (Novus Biological, Immunifluorescence [IF] 1:100) and mouse anti-cytochrome c (BD PharMingen; IF: 1:100) were used to study LIPT2 localization. The amount of co-localization between two channels was quantified using thresholded Pearson’s correlation coefficients with JACoP28 plugin from NIH FIJI software (v.1.51a). To quantify mitochondrial areas and the amount of co-localization at peripheral sites, threshold Pearson’s coefficients were calculated in three randomly chosen 100 pixel × 100 pixel squares in the cell periphery. For averaged line scans, line profiles were calculated as the mean fluorescence intensity averaged over 100 pixels. Maximum intensity projections were calculated from z stacks with 500 nm spacing between slices covering the whole cell. For analysis of immunocytochemistry experiments, a minimum of three independent experiments was performed and statistically significant estimates for each sample were obtained by choosing an appropriate sample size, correlating to 15–40 images per condition per experiment for microscopy-based quantifications. Cells were chosen arbitrarily according to the fluorescent signal in a separate channel, which was not used for quantification. Data are presented as mean ± standard deviation. Statistical tests were performed using a two-tailed, unpaired t test, without excluding samples from statistical analysis. Images were computed using the plugin FigureJ (Jerome Muitterer and Edda Zinck; v.1.10b) on FIJI software.