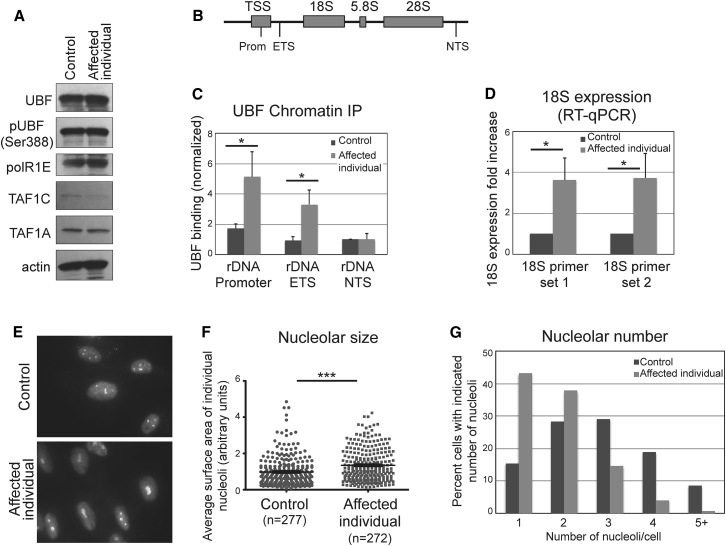
Figure 3.
Mutant UBF Is Hyperactive and Drives Increased Expression of rDNA Genes, Associated with Nucleolar Alterations
(A) Western blot analysis of cells of control and affected individual showing that the UBTF mutation does not affect UBF steady state levels, its phosphorylation status, or the levels of the Pol I subunits PolR1E, TAF1A, and TAFC.
(B) Schematic representation of the rDNA gene locus, indicating the location of each primer set used: Promoter, external transcribed spacer (ETS), and non-transcribed spacer (NTS).
(C) UBF ChIP-qPCR showing that UBF binding to the Promoter and ETS region is significantly increased in cells of the affected individual compared to control cells. Binding was quantified relative to input material, and shown as normalized to the NTS locus in control cells. Bars represent the average of three independent experiments, and error bars represent SD. The asterisk indicates statistical significance (p < 0.05) evaluated using the t test (two-tailed, equal variance).
(D) Quantification of 18S expression in cells of affected individual and control, using real-time qPCR, quantified relative to GAPDH expression. Two different 18S primer sets were used. The results are shown as normalized to control cells. Bars represent the average of three independent experiments, and error bars represent SD. The asterisk indicates statistical significance (p < 0.05) evaluated using the t test (two-tailed, equal variance).
(E) Representative micrographs of anti-nucleolin immunofluorescence showing altered nucloelar pattern in affected individual cells.
(F) Quantification of nucleolar size. Nucleolar area was quantified using ImageJ and statistical analysis was done with Prism (t test, two-tailed, equal variance). Asterisks indicate statistical significance (p < 0.0001). The mean ± SEM is also shown.
(G) Quantification of the number of nucleoli in each cell (117 control and 151 affected individual cells were analyzed).