Figure 2.
Characterization of MRPS34 in Affected Subjects with MRPS34 Mutations
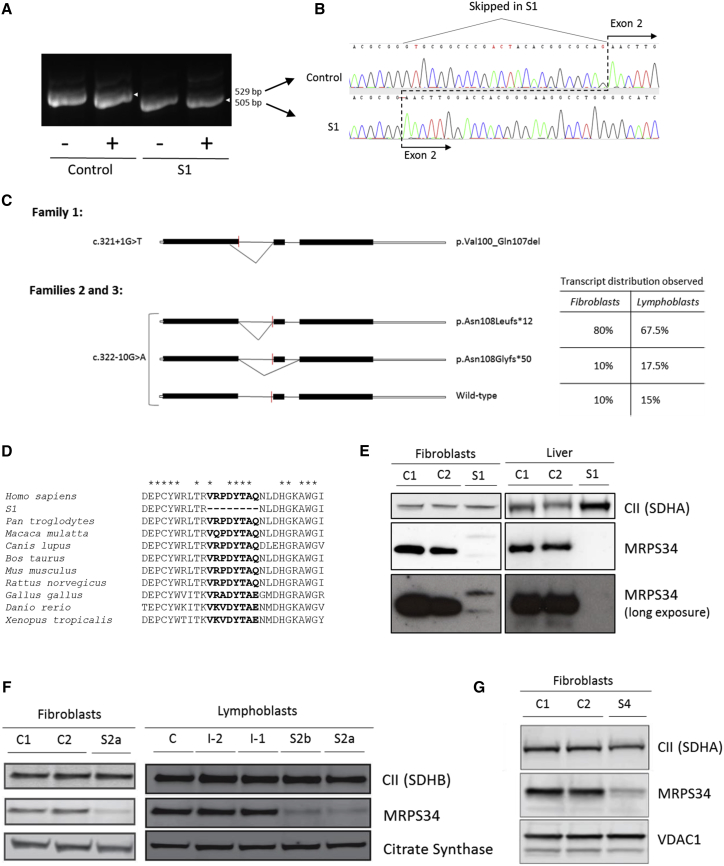
(A) PCR amplicons of MRPS34 exons 1–3 generated from control individual and subject 1 fibroblast cDNA ± cycloheximide (CHX). The amplicon detected in subject 1 was smaller than the control amplicon.
(B) Sequence analysis of the MRPS34 cDNA PCR amplicon detected in subject 1 identified a 24 bp deletion corresponding to the utilization of an upstream donor site in exon 1. This splicing mutation therefore produces a shortened transcript that results in an in-frame deletion of eight amino acids, p.Val100_Gln107del.
(C) Schematic diagram depicting the abnormal transcript generated from the c.312+1G>T variant (family 1) and two abnormal plus residual wild-type transcript generated from the c.322−10G>A variant (families 2 and 3). The distribution of the three transcripts generated from the c.322−10G>A variant in S2a fibroblasts (40 clones sequenced) and lymphoblasts (40 clones sequenced) is additionally described in the table. The red line indicates the position of the variant. The diagram solid black bars represent exons, while the open bars represent untranslated region.
(D) Protein sequence alignment of human MRPS34 with its homologs in nine other vertebrate species. Asterisks (∗) depict conserved amino acids. The eight amino acids missing from the MRPS34 protein produced in subject 1 are highly conserved across the species examined.
(E) SDS-PAGE western blot of MRPS34 and complex II 70 kDa subunit SDHA (loading control) from control individuals (C1 and C2) and subject 1 fibroblasts and liver showed the absence of wild-type MRPS34 protein in subject 1. Long exposures revealed faint double banding in subject 1 fibroblast samples probed with MRPS34 antibody.
(F and G) SDS-PAGE western blot of MRPS34 in fibroblasts and lymphoblasts revealed a substantial decrease in MRPS34 levels in subjects 2a (F), 2b (F), and 4 (G) relative to control individuals (C1 and C2) and to parental samples (I-2 and I-1 from family 2). Complex II subunits SDHA and SDHB, VDAC1, and citrate synthase represent loading controls.