Figure 4. Diversity in the Structures of Various Histone Chaperones.
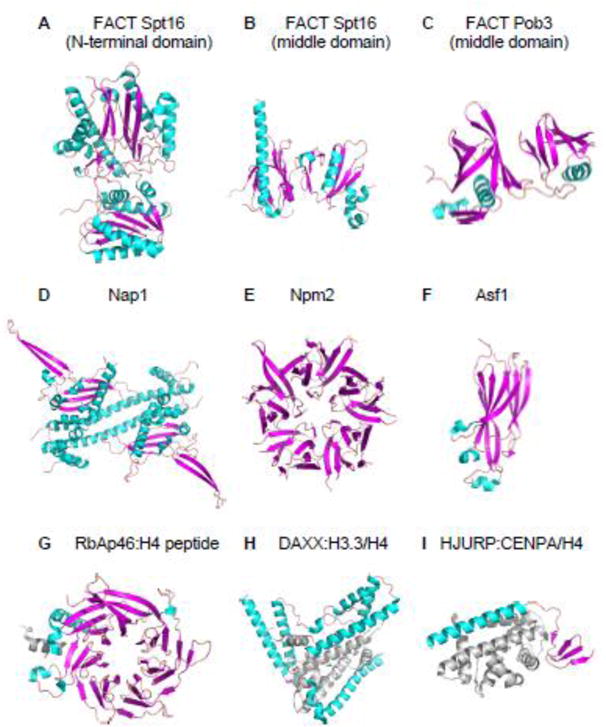
A. Crystal structure of the N-terminal domain (residues 1–451) of yeast Spt16 subunit of FACT (PDB: 3BIT). Residues 452–1035 are not included. Helices, sheets and loops of histone chaperones are colored cyan, magenta, and light pink, respectively, throughout this figure.
B. Crystal structure of the middle domain (residues 653–944) of yeast Spt16 subunit of FACT (PDB: 4KHO). Residues 1–652 and 945–1035 are not included.
C. Crystal structure of the middle domain (residues 237–474) of yeast Pob3 (homologue of human SSRP1) subunit of FACT (PDB: 2GCL). Residues 1–236 and 474–552 are not included.
D. Crystal structure of the core domain (residues 74–365) of yeast Nap1 dimer (PDB: 2Z2R). Residues 1–73 and 366–417 are not included.
E. Crystal structure of the core domain (residues 16–118) of Xenopus laevis Nucleoplasmin pentamer (PDB: 1K5J). Residues 1–15 and 119–195 are not included.
F. NMR structure of the core domain (residues 1–156) of human Asf1a (PDB: 1TEY). Residues 157–204 are not included.
G. Crystal structure of human RbAp46 (residues 7–410) in complex with H4 peptide (residues 28–42) (PDB: 3CFV). Residues 1–6 and 411–425 of RbAp46 are not included. H4 peptide colored gray.
H. Crystal structure of the core domain (residues 181–387) of human DAXX in complex with a H3.3/H4 dimer (PDB: 4HGA). Residues 1–180 and 388–740 of DAXX are not included. H3.3/H4 dimer colored gray.
I. Crystal structure of human HJURP (residues 14–74) in complex with a CENP-A/H4 dimer (PDB: 3R45). Residues 1–13 and 75–748 of HJURP are not included. CENP-A/H4 dimer colored grey.
