Figure 5. Recent Structural Insights into the Roles of Disordered Regions in Binding Histones.
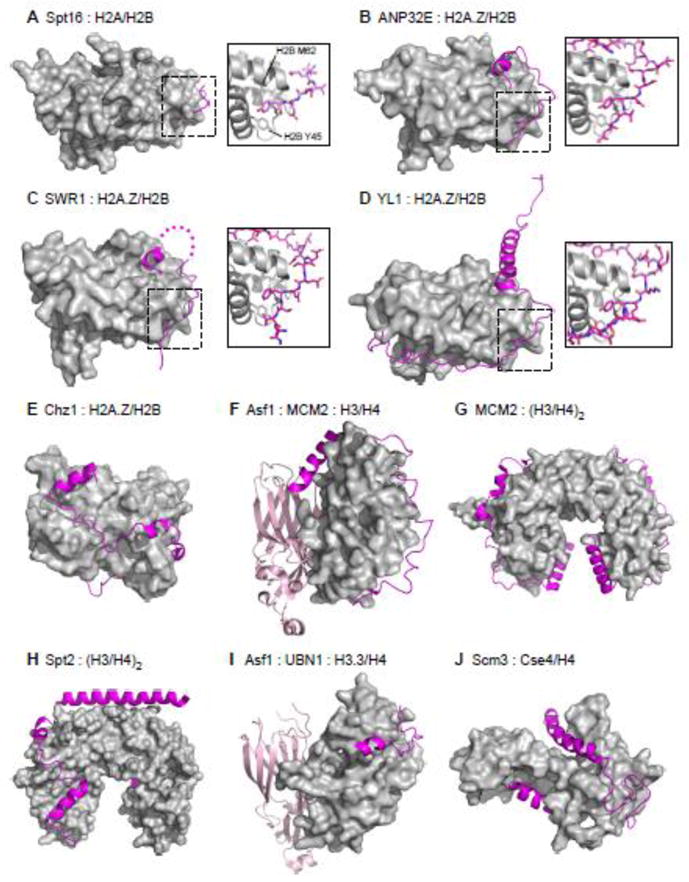
A. Crystal structure of a peptide derived from the acidic disordered C-terminus of yeast Spt16 (residues 967–972) bound to a H2A/H2B dimer (PDB: 4WNN). Chaperone peptide represented as a magenta ribbon, histones represented as grey surface. Boxed: Details of the interaction, with Spt16 peptide represented as magenta sticks and histones shown as gray ribbon with H2B residues Tyr45 and Met62 shown as sticks. Spt16 anchors a conserved tyrosine into the hydrophobic pocket created by H2B Tyr45 and Met62, and uses neighboring acidic residues to cap helix 2 of H2B.
B. Crystal structure of a peptide derived from the acidic disordered C-terminus of human ANP32E (residues 215–237) bound to a H2A.Z/H2B dimer (PDB: 4CAY). Same coloring and representation used as in A. ANP32E also shows conserved “anchoring and capping” binding mechanism to the same site on the H2A.Z/H2B dimer. Additional contacts are made between H2A.Z specific residues and ANP32E, and are necessary for chaperone specificity.
C. Crystal structure of a peptide derived for the Z-domain of yeast SWR1 chromatin remodeler (residues 598–628) bound to a H2A.Z/H2B dimer (PDB: 4M6B). Same coloring and representation used as in A. SWR1 also shows conserved “anchoring and capping” binding mechanism to the same site on the H2A.Z/H2B dimer.
D. Crystal structure of a peptide derived from the acidic disordered N-terminus of Drosophila YL1 histone chaperone component of the SCRAP/SWR1 chromatin remodeling complex (residues 4–71) bound to a H2A.Z/H2B dimer (PDB: 5CHL). Same coloring and representation used as in A. YL1 also shows conserved “anchoring and capping” binding mechanism to the same site on the H2A.Z/H2B dimer.
E. NMR structure of yeast Chz1 histone chaperone core (residues 1–62) bound to a H2A.Z/H2B dimer (PDB: 2JSS). Same coloring and representation used as in A. Chz1 binds to H2A.Z/H2B via a long irregular extended conformation capped by short N and C-terminal helices. No clear anchoring a capping motif is apparent in this structure.
F. Crystal structure of the quaternary complex of human histone chaperone Asf1a core (residues 2–154) and replicative helicase MCM2 (residues 69–121) bound to a H3/H4 dimer (PDB: 5C3I). Same coloring and representation used as in A, but with Asf1 core represented as a light pink ribbon. MCM2 adopts an extended conformation capped by a C-terminal helix. Tyr90 of MCM2 is anchored into a hydrophobic pocket created by H3 helices α1 and α2, with adjacent acidic residues making electrostatic contacts with histone surface residues similar to conserved “anchoring and capping” binding motifs.
G. Crystal structure of human replicative helicase subunit MCM2 (residues 69–121) bound to a H3/H4 tetramer (PDB: 5BNV). Same coloring and representation used as in A. MCM2 adopts a very similar conformation as in F.
H. Crystal structure of the human histone chaperone Spt2 (residues 606–675) bound to a H3/H4 tetramer (PDB: 5BS7). Same coloring and representation used as in A. Spt2 binds to the H3/H4 tetramer in an extended conformation capped by helices, most notably a long acidic C-terminal helix which bridges the dimer-dimer interface in the H3/H4 tetramer. No clear anchoring a capping motif is apparent in this complex structure.
I. Crystal structure of the quaternary complex of yeast histone chaperone Asf1 core (residues 5–154) and a peptide from UBN1 (residues 122–142) bound to a H3.3/H4 dimer (PDB: 4ZBJ). Same coloring and representation used as in A, but with Asf1 core represented as a light pink ribbon. The UBN1 peptide adopts an extended conformation capped by a small N-terminal helix. Tyr132 and Phe138 of UBN1 are anchored into hydrophobic grooves of the H3.3/H4 dimer, and Asp133, Asp136, and Asp140 form electrostatic contacts similar to other anchoring and capping motifs.
J. NMR structure of the Cse4-binding domain of yeast Scm3 (residues 90–169) bound to a Cse4/H4 dimer. Same coloring and representation used as in A. Scm3 binds to Cse4/H4 in an extended conformation with N and C-terminal helices. The N-terminal helix of Scm3 contains residues Trp107, Ile110, Ile111, Tyr114, and I117 that make hydrophobic contacts with many Cse4-specific residues, and many acidic residues on the middle loop and C-terminus likely make electrostatic contacts.
