Figure 4.
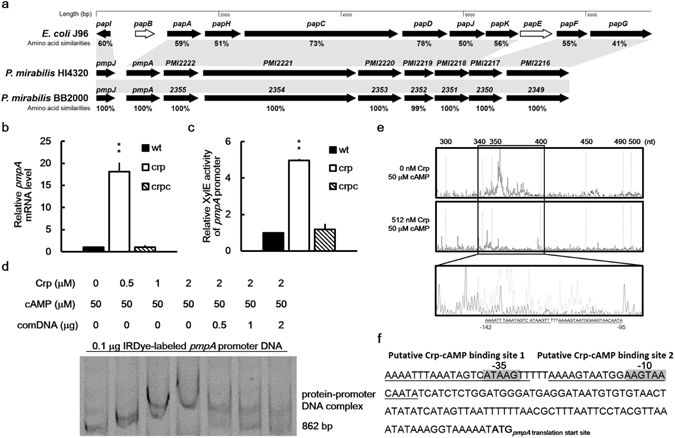
Regulation of pmpA expression by P. mirabilis Crp-cAMP. (a) The P-like fimbrial gene locus (pmp) similar to the well characterized pap locus in UPEC J96 in P. mirabilis genome. An amino acid sequence analysis of the pmp locus in P. mirabilis HI4320 (accession no. AM942759) and its counterparts in E. coli J96 (accession no. ALIN02000070) and P. mirabilis BB2000 (accession no. NC_022000) was performed using position-specific iterative BLAST. The nine proteins of the pmp locus in P. mirabilis HI4320 or BB2000 are similar to the PapI, PapA, PapH, PapC, PapD, PapJ, PapK, PapF and PapG in E. coli J96 with corresponding genes in shadows. The percent amino acid similarities between P. mirabilis HI4320 and E. coli J96 or P. mirabilis BB2000 were shown below each gene. The white arrows represent genes that are not found in either P. mirabilis HI4320 or P. mirabilis BB2000. (b) Loss of crp increased the pmpA mRNA level. The pmpA mRNA levels of the wild-type, crp mutant, and crp-complemented strain were quantified using real-time RT-PCR at 24 h after incubation. The value obtained for the wild-type was set at 1. (c) Loss of crp increased the promoter activity of pmpA. The activities of XylE in the pmpA-xylE reporter plasmid-transformed wild-type, crp mutant, and crp-complemented strain were determined using the reporter assay at 7 h after incubation. The value obtained for the wild-type was set at 1. In (b and c), the data represent the averages and standard deviations of three independent experiments. The significant difference from the wild-type was determined by Student’s t test (**P < 0.01). wt, wild-type; crp, crp mutant; crpc, crp-complemented strain. (d) The binding of P. mirabilis Crp-cAMP to pmpA promoter region revealed using an EMSA. The EMSA was performed as described in Fig. 3d, except that IRDye-labeled pmpA promoter region DNA fragments (862 bp) were incubated with 0–2 μM of the purified His-tagged Crp. (e) Identification of the Crp-cAMP binding site in the pmpA promoter region by a DNase I footprinting assay. FAM was used to label the pmpA promoter DNA fragment and the DNA fragment was incubated with cAMP (50 μM) with or without the recombinant Crp (512 nM) followed by DNase I treatment. The mixture was subject to electrophoresis. The fluorescence intensity of the FAM-labeled DNA fragment (ordinate) was plotted against the sequence length of the fragment. Two Crp-cAMP binding sites (underlined) located between −95 and −142 upstream of pmpA start codon were shown in an expanded view. (f) A diagram showing the Crp-cAMP binding sites upstream of pmpA gene. The putative Crp-cAMP binding sequences are underlined and the putative −10 and −35 promoter sequences of sigma 70 are shadowed. The full-length gel for (d) is shown in Supplementary Fig. S8.
