Fig. 2.
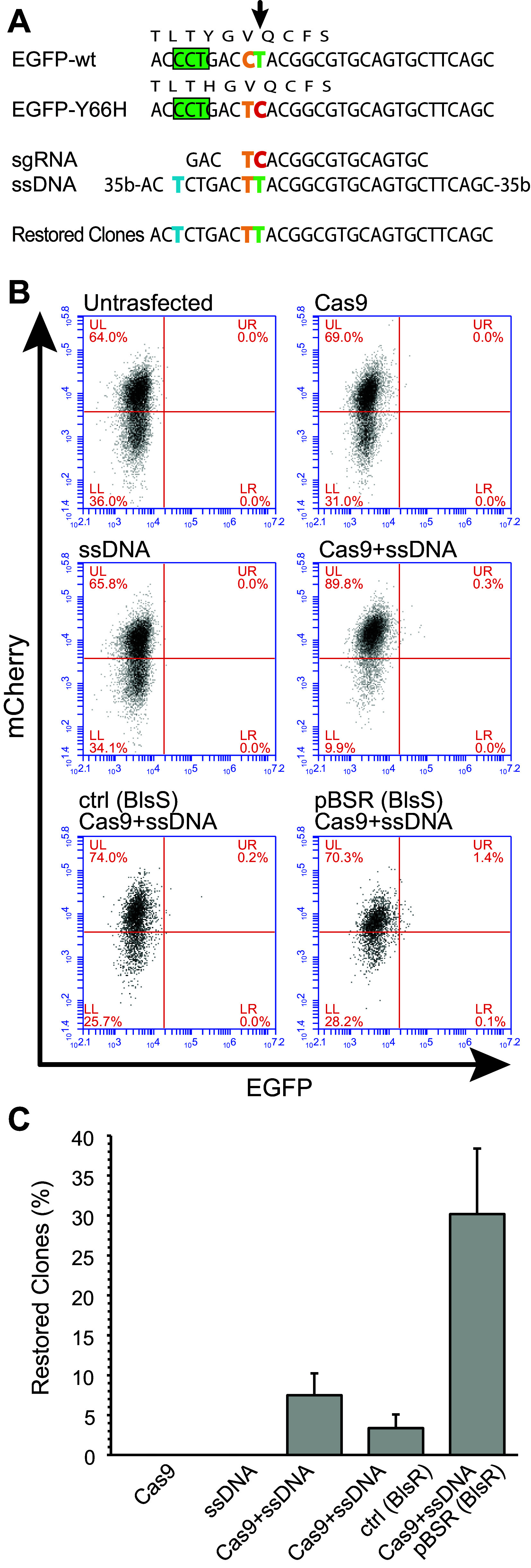
Reversion of an inactive EGFP reporter gene by knock-in. a A single point mutation (T>C) (green > red, black arrow) disables the EGFP in an mCherry-EGFPY66H reporter cassette stably transfected in a HEK293T clone. To restore the EGFP fluorescence, an sgRNA for the mutated sequence (PAM sequence indicated by the green box) was used in combination with a single-stranded 100nt DNA fragment (ssDNA) bearing both the restoring mutation (green) and a mutated PAM sequence (blue), to escape cleavage by Cas9. A C>T polymorphism (yellow) was also inserted to distinguish the sequence in mutated clones from potential contamination by wild-type EGFP. Sequencing of clones selected with our enrichment system (Restored) confirmed the knock-in of the restoring mutation. b Representative FACS analysis of green fluorescence restoration in mCherry-EGFPY66H expressing cells using either sgRNA/Cas9 alone (Cas9), ssDNA, sgRNA/Cas9 coupled with the ssDNA fragment (Cas9 + ssDNA), or the sgRNA/Cas9 and the ssDNA fragment together with the pBSR plasmid (pBSR(BlsR) + Cas9 + ssDNA) or with a control plasmid (ctrl(BlsR) + Cas9 + ssDNA). mCherry-EGFPY66H stable clones were transiently transfected. The cells were analysed by FACS 1 week after antibiotic selection of the pBSR-transfected samples. The percentage of cells that reacquire the green fluorescence is indicated in the upper right quadrant (UR). At least three independent experiments were performed (average population in the UR quadrant: 0% for untransfected, ssDNA, and Cas9; 0.2% for Cas9 + ssDNA; 0.1% for ctrl(BlsR) + Cas9 + ssDNA; 1.1% for pBSR(BlsR) + Cas9 + ssDNA). (c) Efficiency of knock-in in clones obtained using the different protocols. The bar diagram indicates the percentage of clones in which the EGFP fluorescence was restored (overall number of clones: Cas9, 45; ssDNA, 42; Cas9 + ssDNA, 131; ctrl(BlsR) + Cas9 + ssDNA, 39; pBSR(BlsR) + Cas9 + ssDNA, 70). The error bars indicate the SEM from at least three independent experiments. Enrichment of EGFP-restored clones using the pBSR is significantly different from all treatments (p < 0.05)
