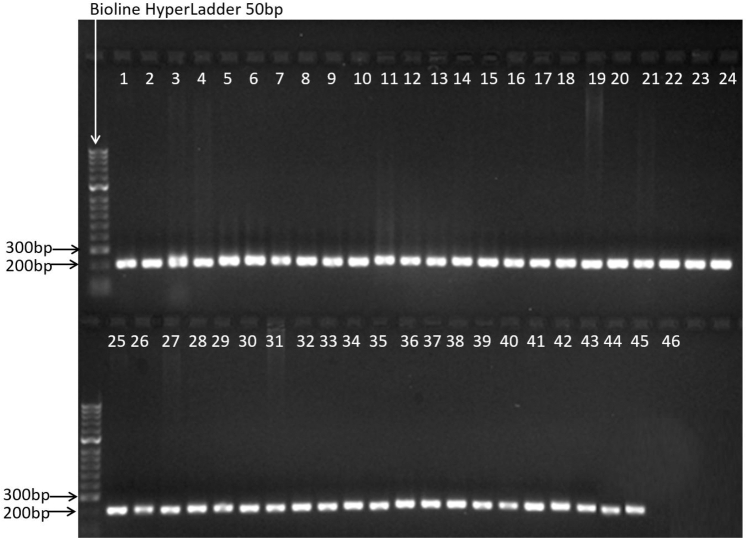
Figure 1:
Gel showing amplification success from 45 known tissue samples representing 40 species, using the AusPreda_12S mini-barcode primers developed in this study, and a PCR negative. The expected amplicon size is 218 bp. Samples are grouped by species as follows: lanes 1 and 2: Felis catus; 3: Canis lupus familiaris; 4: Canis lupus dingo; 5 and 6: Dasyurus viverrinus; 7 and 8: Dasyurus maculatus; 9 and 10: Vulpes vulpes; 11 and 12: Sarcophilus harrisii; 13: Oryctolagus cuniculus; 14: Lepus capensis; 15: Bos Taurus; 16: Ornithorhyncus anatinus; 17: Trichosorus vulpecula; 18: Petaurus breviceps; 19: Tachyglossus aculeatus; 20: Potorous tridactylus; 21: Bettongia gaimardi; 22: Dactylopsila trivirgata; 23: Burramys parvus; 24: Macropus rufogriseus; 25: Thylogale billardierii; 26: Pseudomys gracilacaudatus; 27: Pseudocheirus peregrinus; 28: Antechinus minimus; 29: Tiliqua nigrolutea; 30: Vombatus ursinus; 31: Isoodon obesulus; 32: Macropus giganteus; 33: Parameles gunnii; 34: Sminthopsis leucopus; 35: Mus musculus; 36: Planigale gilesi; 37: Rattus lutreolus velutinus; 38: Phascogale tapoatafa; 39: Hydromys chrysogaster; 40: Macropus rufus; 41: Vicugna pacos; 42: Dasyurus hallucatus; 43: Lathamus discolour; 44: Geocrinia laevis; 45: Dasyurus geoffroii; 46: PCR negative.