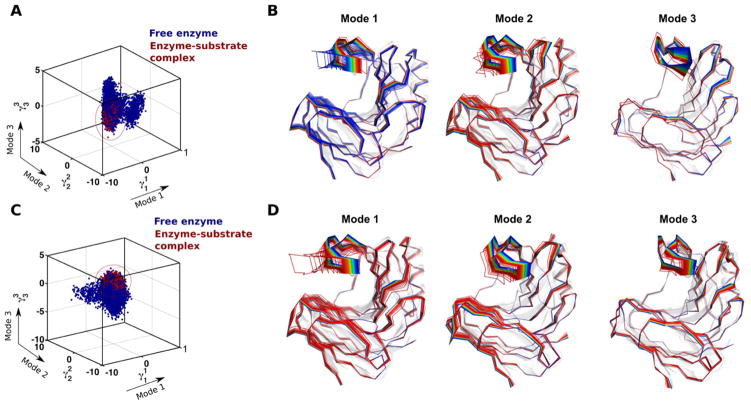
Figure 4. Conformational sub-states of xylanase B2 (XlnB2) from Streptomyces lividans determined using computational simulations and QAA.
Representative conformations along the top three modes for the interconversion between free and (A–B) X6-bound and (C–D) X9-bound XlnB2 binary complexes. A total of 200,000 conformational snapshots obtained from the MD simulations were used as input for QAA to identify the top QAA-independent component vectors for characterizing the primary dynamics associated with the substrate binding process in XlnB2. Reprinted with permission from ref. 82. Copyright 2016 American Chemical Society.