Fig. 4.
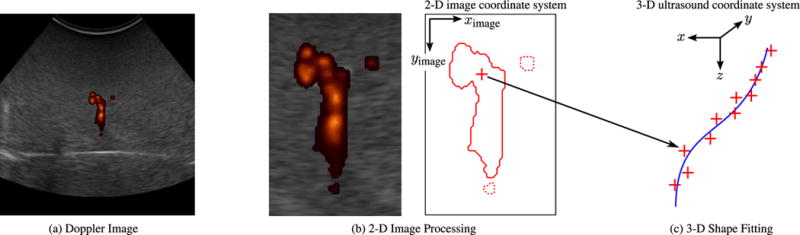
Segmentation algorithm: (a) Doppler image. A 2-D ultrasound Doppler image from a series of N captured over a sweep of the imaging plane. (b) 2-D image processing. Doppler patches with fewer than 300 connected pixels are assumed to be noise and discarded (dotted lines). The remaining Doppler data (solid line) are used to estimate the position of the needle cross section (cross). In the lateral (ximage) direction, the position is estimated as the centroid of the Doppler response. In the axial (yimage) direction, the position is estimated as the lower bound of the region that contains one quarter of the integrated Doppler intensity. (c) 3-D shape fitting. The 2-D-localized points are combined in 3-D based on position information from the 3-D ultrasound transducer. Third-order polynomial curves are fit to the identified points in order to define the needle’s 3-D shape over the series of N images.
