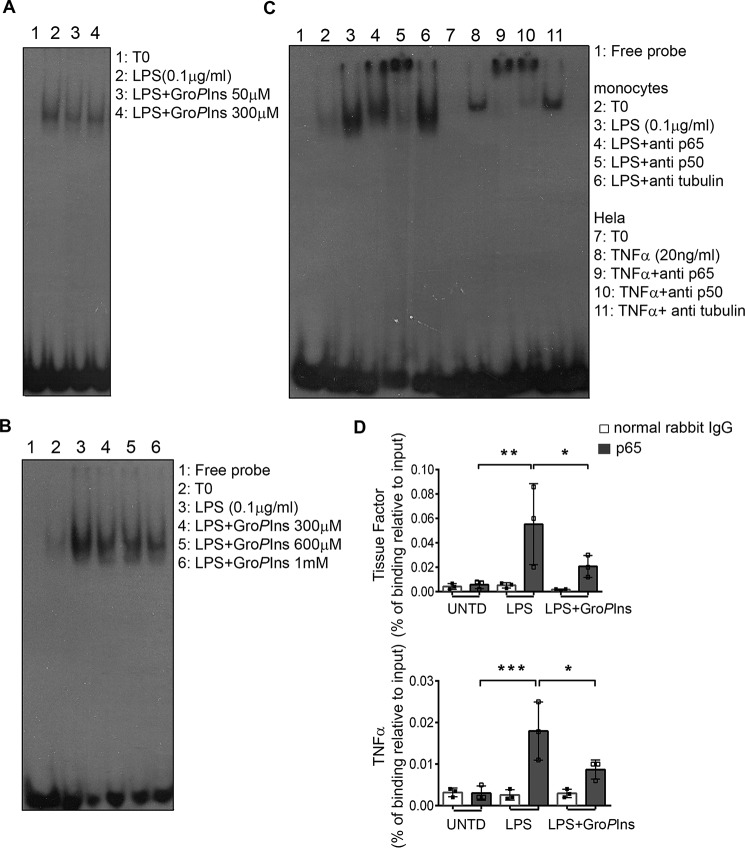
Figure 5.
GroPIns reduces the LPS-induced binding of NF-κB to promoters of target genes. A, GroPIns reduces the LPS-triggered binding of NF-κB to DNA. For the EMSA, monocytes were incubated at 37 °C for 20 min without or with the indicated concentrations of GroPIns and then incubated for a further 30 min in the presence of 0.1 μg/ml LPS. Unstimulated monocytes were incubated in parallel as a control (T0). At the end of the incubation, the cells were lysed, and the nuclear extracts were incubated with radiolabeled oligonucleotide probes that contained the NF-κB-binding site (see “Experimental procedures”). Protein–DNA complexes were separated using 5% non-denaturing acrylamide gels and visualized by autoradiography. Data are representative of two independent experiments. B, GroPIns directly displaces NF-κB from DNA. For the EMSA competition assay, GroPIns (at the indicated concentrations) was added to the binding mixture containing the radiolabeled probe and the nuclear lysates from monocytes treated with 0.1 μg/ml LPS for 60 min. The EMSA was performed as reported above. Data are representative of three independent experiments. C, left side of the blot, both p65–p50 heterodimer and p50–p50 homodimer participate in the binding to the radiolabeled probe. For the EMSA supershift, antibody specific for p65 or p50 protein was added to the binding mixture containing the radiolabeled probe and the nuclear lysates from monocytes treated with 0.1 μg/ml LPS for 60 min; an antibody specific for tubulin was used as a negative control. C, right side of the blot, HeLa cells treated with 20 ng/ml TNF-α for 20 min were used as a positive control. The EMSA was performed as reported above. D, GroPIns reduces the recruitment of p65 to promoters of tissue factor and TNF-α genes. For the ChIP assay, the binding of p65 on chromatin was evaluated by a specific rabbit polyclonal antibody to p65 protein; a normal rabbit IgG was used as a background control. The enriched promoters were quantified by real-time PCR using primers that specifically amplify a region containing the κB site. The intensity of the PCR signal is proportional to the occupancy on the binding site. The amount of p65-immunoprecipitated chromatin is represented as signal relative to the total amount of input chromatin (percentage of binding relative to input). Data are mean ± S.D. of three independent experiments with cells from three independent donors. Significance was tested by analysis of variance with Dunnett post hoc analysis (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars represent S.D. UNTD, untreated.