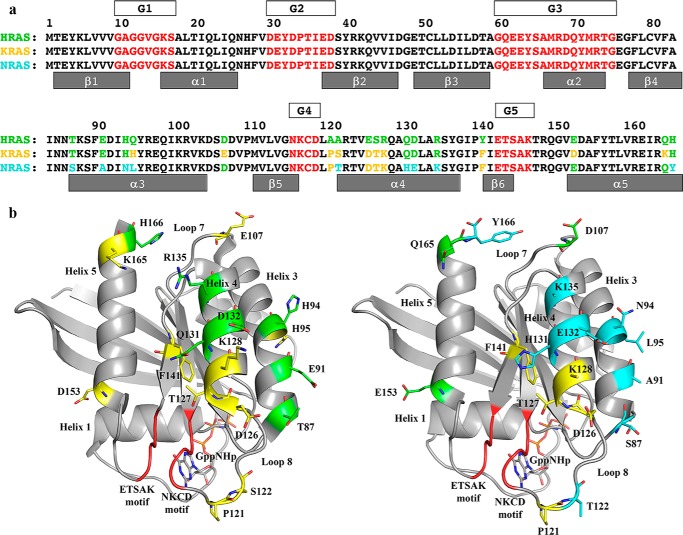
Figure 3.
Amino acid sequence differences between H-, K-, and N-Ras. a, sequences of the three Ras isoforms are shown, numbered every 10 residues. Amino acid residues common to H-, K-, and N-Ras are in black. Residues that are different in any isoform are shown in green in the H-Ras sequence (top row). Residues in K-Ras that are the same as in H-Ras are shown in green in the K-Ras sequence, and those that are different from H-Ras are shown in yellow (middle row). Residues in N-Ras that are the same as in H-Ras are shown in green, and those that are the same as in K-Ras are shown in yellow, and those that are unique to N-Ras are in cyan (bottom row). The G elements found in all GTPases are shown in red with boxes above the Ras sequences labeled G1–G5. In the effector lobe, G1 is the P-loop; G2 is switch I; and G3 is switch II. In the allosteric lobe, G4 and G5 are the NKXD and EXSAK motifs, respectively, important for binding the guanine base of the nucleotide. b, amino acid differences mapped onto the three-dimensional structure of Ras. Left, K-Ras structure (PDB code 3GFT) with color scheme as shown for the sequence of K-Ras in a. Right, N-Ras structure (PDB code 5UHV, presented here) with color scheme as shown for the sequence of N-Ras in a.