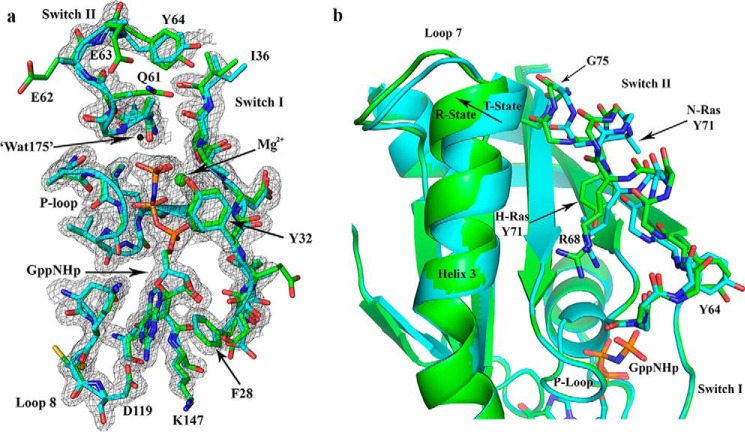
Figure 4.
Comparison between N-Ras and H-Ras bound to GppNHp, both solved from crystals with symmetry P3221. a, active site shown with 2Fo − Fc electron density map for the N-Ras structure, contoured at the 1σ level. The model for N-Ras (PDB code 5UHV, presented here) is shown in cyan and that for H-Ras (PDB code 1CTQ) is in green. The three phosphorus atoms of GppNHp are shown in orange, and the nitrogen atom that bridges the β- and γ-phosphates is in blue. The nucleotide-binding residues are shown, including the P-loop, switch I, switch II, and NKXD and EXSAK motifs. The magnesium ion is shown as a green sphere, and the nucleophilic water molecule found in the H-Ras structure (Wat-175) is shown as a black sphere. b, T state to R state conformational differences are indicated with an arrow at the C-terminal end of helix 3. Differences are also found in loop 7 and switch II. Disordered side chains were omitted from the model. For example, the N-Ras Tyr-71 side chain is represented up to the Cβ atom. The superposition of N-Ras and H-Ras was based on the nucleotide.