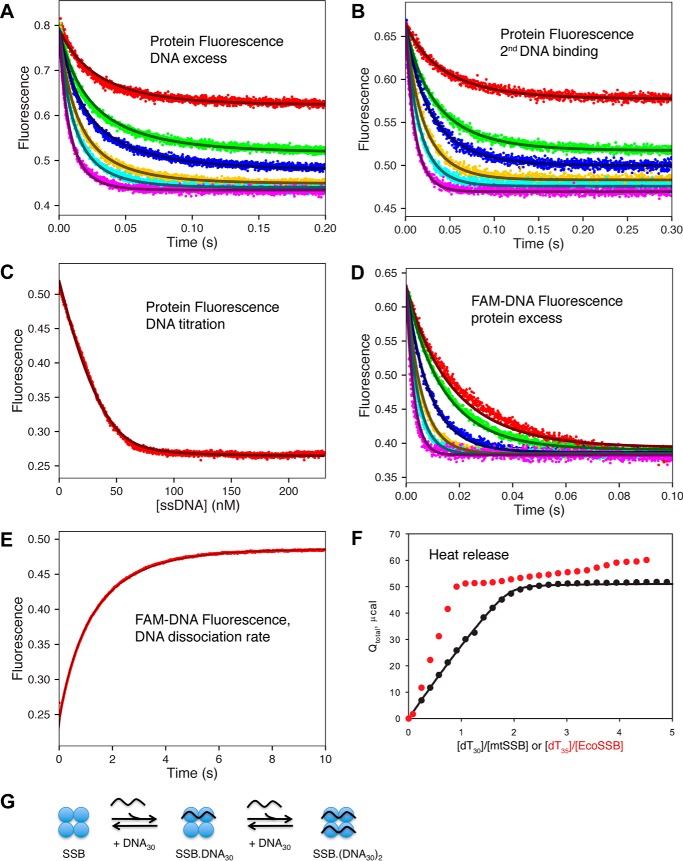
Figure 4.
Kinetics, equilibria, and thermodynamics of mtSSB binding to dT30 ssDNA. A, the kinetics of mtSSB binding to dT30 ssDNA were monitored by protein fluorescence with DNA in excess (except for the lowest concentration). [DNA] = 15, 30, 40, 60, 80, and 120 nm; [mtSSB] = 20 nm. B, kinetics of binding a second ssDNA to mtSSB–DNA30. Equimolar concentrations of mtSSB and dT30 (42 nm each) were allowed to equilibrate and then were diluted 1:1 with various concentrations of dT30 (15, 30, 40, 60, 80, and 120 nm, final concentrations), and the protein fluorescence was recorded. C, equilibrium titration of mtSSB (30 nm) with increasing concentrations of dT30 monitored by following protein fluorescence. D, the kinetics of mtSSB binding to FAM-dT30 were monitored by fluorescein fluorescence with protein in excess. [FAM-dT30] = 20 nm; [mtSSB] = 40, 50, 80, 120, 160, and 240 nm. E, measurement of the dissociation rate of FAM-dT30 from mtSSB. The fluorescein-labeled DNA fluorescence was monitored upon mixing a preformed 1:1 mtSSB–dT30 complex (20 and 22 nm, respectively) with an excess of unlabeled dT60 (600 nm). F, measurement of heat release (μcal) upon titration of mtSSB (490 nm) with increasing concentrations of dT30 (0–2,500 nm), as measured by ITC. Data for ITC titration of EcoSSB with dT35 (red dots) are shown as a comparison, with an x axis of dT35/EcoSSB. All data for mtSSB kinetics and equilibria were fit globally to the model shown in Scheme 1 to give the smooth lines based upon numerical integration of the rate equations with no simplifying assumptions. Fluorescence is given in arbitrary units, and in global data fitting, individual experimental outputs were modeled with different scaling factors to reflect the setup of the instrument on different days. G, schematic showing the sequential binding of two dT30 strands of ssDNA to the mtSSB tetramer (SSB). Note that for EcoSSB, the binding of the second DNA is much weaker (negative cooperativity), which is not apparent with mtSSB.