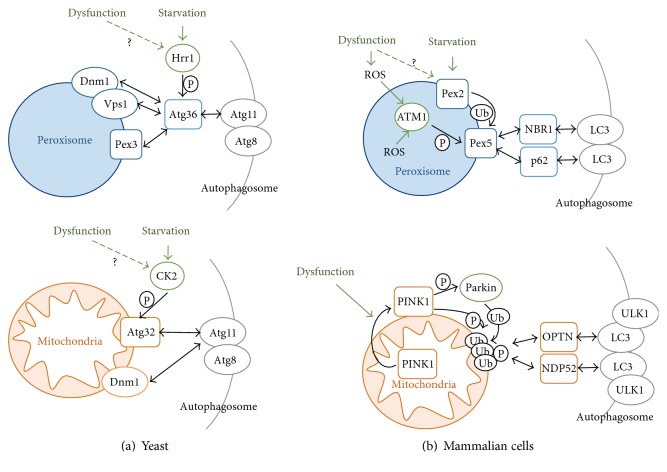
Figure 2.
Mechanisms of autophagic removal of peroxisomes and mitochondria. In the upper panel, pexophagy mechanisms are depicted for budding yeast (a) and mammalian cells (b). In yeast, pexophagy is induced by the Hrr1 kinase which phosphorylates the Atg36 adaptor. Atg36 contacts the Pex3 peroxisomal receptor, the fission machinery (Dnm1, Vps1), and the autophagosomal adaptor Atg11. In mammalian cells, a dysfunctional peroxisome and general ROS increase activate the ATM kinase, which phoshorylates the Pex5 receptor. Pex5 is additionally targeted by ubiquitination via the starvation-inducible Pex2. Modified Pex5 interacts with the autophagosomal adaptors NBR1 and p62. In the lower panel, mitophagy mechanisms are depicted for budding yeast (left) and mammalian cells (right). In yeast, starvation induces mitophagy via casein kinase 2 (CK2), which phosphorylates the Atg32 receptor. Modified Atg32 interacts with the Atg11 autophagosomal adaptor, which also contacts the mitochondrial fission machinery (Dnm1). In mammalian cells, mitochondrial dysfunction triggers the exposure of PINK1 at the organelle surface. PINK1 phosphorylates and activates the Parkin ubiquitin ligase, which marks outer mitochondrial membrane proteins. PINK1 additionally phosphorylates polyubiquitin chains at mitochondria, which leads to recognition by the autophagosomal adaptor proteins Optineurin (OPTN) and NDP52. P = phosphorylation; Ub = ubiquitination.