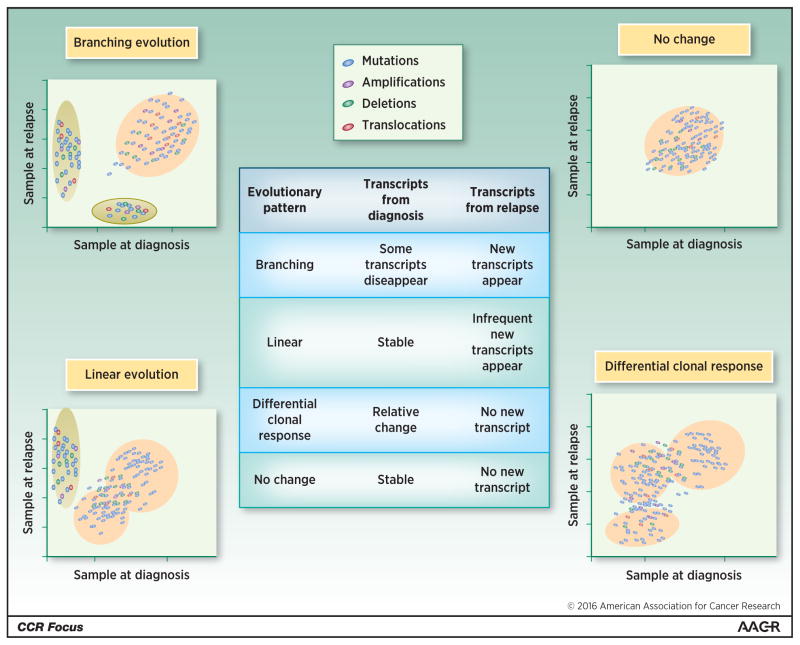
Figure 1. Clonal heterogeneity and clonal evolution in multiple myeloma impacts gene expression profile.
The figure represents complex clonal content and clonal evolution in multiple myeloma. Four distinct evolutionary patterns have been identified and their impact on gene expression profile is presented. Branching evolution, which corresponds to the appearance of new clones expressing new genes while other clones disappear, affects GEP. Linear evolution is characterized by the emergence of one new clone that influences GEP. Differential clone response, featured by the modification of the relative proportion of the sub-clones, affects GEP. Stable clonality with no detectable change does not impact GEP.