Figure 5. Deletion analysis of the NOX4 promoter reveals TGFβ/SMAD3 and p53 regulatory sequences.
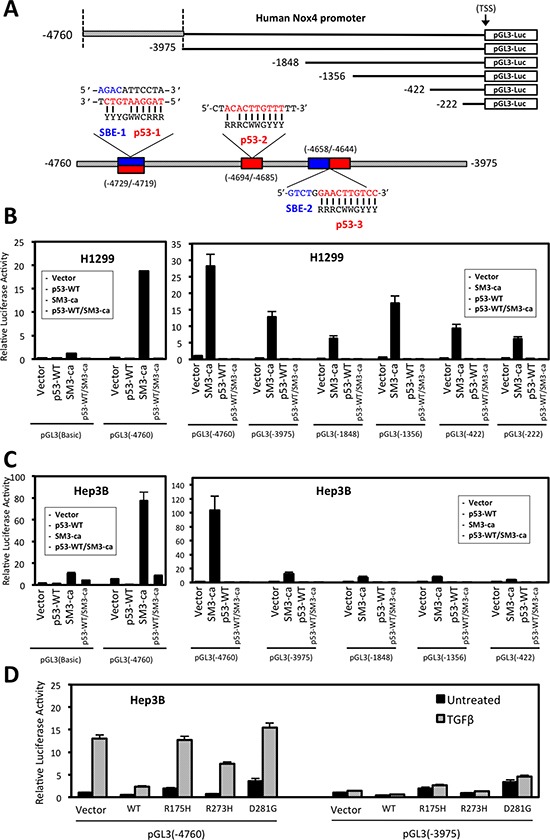
(A) Schematic of 5′-end deletion series of the human NOX4 promoter starting from the transcription start site (TSS) was cloned into the pGL3-luciferase reporter vector to measure promoter activity. Putative SMAD binding elements (SBE) and p53 response elements are indicated within the -4760/-3975 promoter regions. The traditional p53 response element (p53-RE) consensus sequence consists of two 10-base decamers with a 0-13-base spacer: RRRCWWGYYY…n = 0-13 bp…RRRCWWGYYY (R is a purine (A/G), Y is a pyrimidine (C/T), and W is (A/T) [27]. However, studies have shown that p53 target genes that have one of the two 10-base decamers, or “half site” within the promoter can still be regulated by p53 binding [26, 27]. The consensus sequences for SMAD binding elements (SBE) are CAGA and its reverse complement GTCT.[41] Above SBE are indicated in blue and p53-RE in red. (B) H1299 cells and (C) Hep3B cells were co-transfected with designated pGL3-Basic control or NOX4 promoter reporter plasmids and either vector control, SMAD3ca, p53-WT, or both p53-WT/SMAD3ca plasmids. Luciferase activity was determined 48 hours after transfection (n = 3, in triplicate). (D) Hep3B cells co-transfected with NOX4 promoter pGL3 (-4760) or pGL3 (-3975) and either vector control, p53-WT, p53-R175H, p53-R273H, or p53-D281G plasmids for 24 hours. The cells were then left untreated or treated with TGFβ (5 ng/ml) for an additional 24 hours. Total cell lysates were collected following TGFβ treatment and analyzed for luciferase activity (n = 3, in triplicate).
