Figure 7. Defined ERG target gene signature is not increased in 1,25D(OH)2D3-treated VCaP cells.
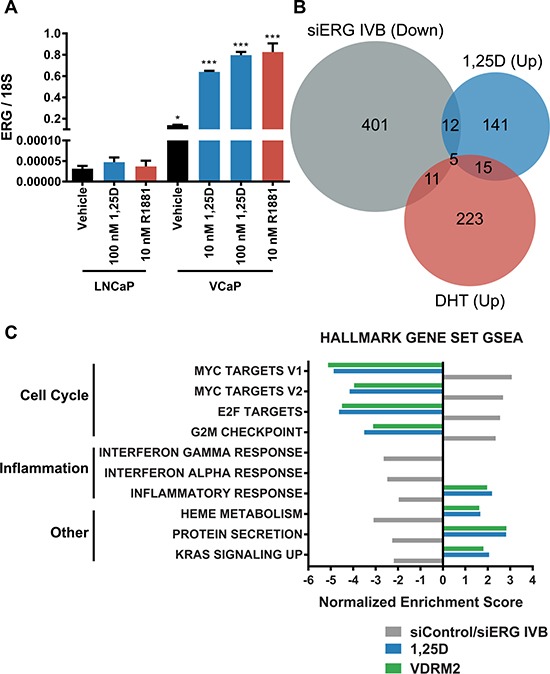
(A) LNCaP or VCaP cells were changed to stripped serum and treated for 24 hours with vehicle (EtOH), 10 nM 1,25D(OH)2D3 (1,25D), 100 nM 1,25D(OH)2D3, or 10 nM R1881 and RNA was purified. ERG mRNA was measured by RT-qPCR and normalized to 18S. (B) Venn diagram showing comparison of a previously published microarray from VCaP cells treated with siRNA against ERG (siERG IVB), a previously published microarray from VCaP cells treated with DHT, and our RNA-seq data from VCaP cells treated with 10 nM 1,25D(OH)2D3. The siERG IVB group denotes genes significantly downregulated at least 1.5-fold relative to control siRNA (genes requiring ERG for expression) and the DHT and 1,25D(OH)2D3 groups denote genes significantly upregulated at least 1.5-fold relative to vehicle control. (C) The top 10 pathways significantly regulated via GSEA using the Hallmark gene set collection comparing siERG IVB (ERG induced),1,25D(OH)2D3, and VDRM2-treated VCaP cells plotted as normalized enrichment score. *p < 0.05, ***p < 0.001, relative to respective LNCaP treatment. n = 3, representative graph, mean ± SEM.
