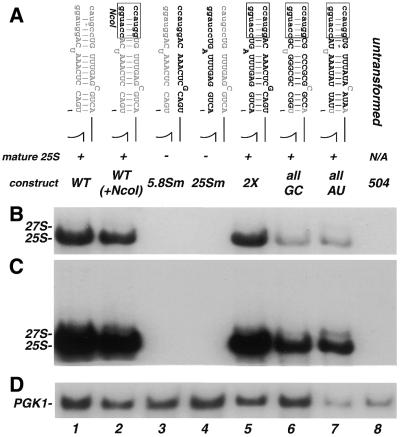
Figure 2.
Analysis of plasmid-encoded rRNA containing mutations within the ITS2-proximal stem. (A) Schematic representation of the mutations and the name by which they are referred to in the text. Wild-type sequences are indicated by gray lettering, mutated sequences are black and the NcoI site is denoted by lower case black letters. Maturation of rRNA scored as (+) normal processing with accumulation of mature 25S; and (–) low or no detectable levels of mature 25S. Below, northern blot analysis of mutations illustrated in (A). Total RNA (8 µg) from each transformant was resolved on a denaturing 1.2% agarose–formaldehyde gel and transferred to nylon membrane. The membrane was divided in half and probed with 5′-end-labeled oligonucleotides complementary to either the tag sequence within 25S (B and C) or PGK1 (D) as a control for loading. (C) Longer exposure of (B) presented to visualize the lower amounts of precursors and mature 25S present in some of the mutants. The positions of mature 25S rRNA precursors are noted on the left. 504 has no plasmid and indicates background hybridization for the tag probe.