Figure 1. Multiplexed-FISH reveals cytogenetic subclonal heterogeneity in CLL.
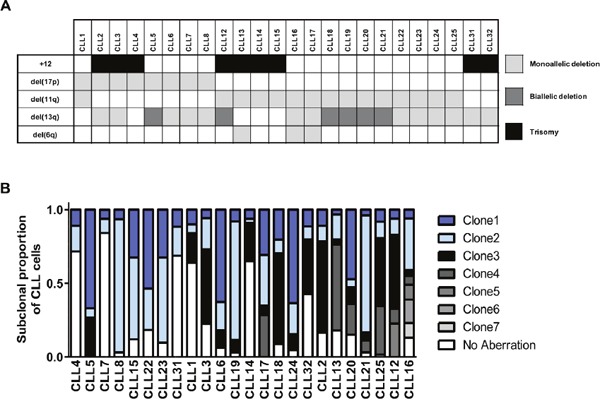
(A) From 128 CLL patients, these 24 pre-treatment CLL samples were identified as amenable for multiplexed-FISH based on the presence of del(6q), del(11q), del(13q), del(17p) and trisomy 12 in various combinations. (B) Multiplexed-FISH with patient-specific probe combinations revealed the cytogenetic architecture of each of the 24 CLLs. The size of each clone is expressed as a proportion of the total number (≥200) of cells analyzed. The samples are arranged according to the cytogenetic complexity, with those having fewer different subclones placed on the left side of the figure. The most complex subclones are presented at the top of each bar. +12 indicates trisomy 12.
