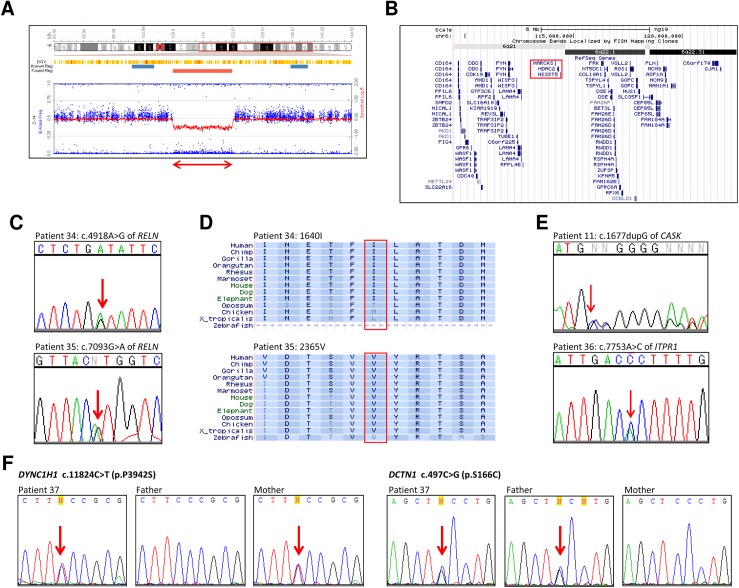
Fig 3. Genomic analysis of candidate genes other than CASK.
(A) Result of the SNP array in patient 33 showing Heterozygous deletion at 6q21-q22.31 including HDAC2 and MARCKS. This result is described as follows: arr 6q21q22.31(109,497,085–122,505,593)x1. The double-headed arrow indicates the deletion. (B) Mapping of the heterozygous deletion in patient 33. The red box denotes HDAC2 and MARCKS. (C) Electropherograms depicting the mutations of RELN detected by targeted resequencing. Arrows indicate the mutated nucleotides. (upper) c.4918A>G (p.I1640V) in patient 34, (lower) c.7093G>A (p.V2365M) in patient 35. (D) Conservation of amino acids around each mutation of RELN in patient 34 (upper) and patient 35 (lower). The red box denotes the amino acid substituted by the mutation. (E) Electropherograms depicting the mutations detected by whole exome sequencing. Each arrow indicates the mutated nucleotide. (upper) c.1677dupG (p.R560Afs*20) of CASK in patient 11, (lower) c.7753A>C (p.T2585P) of ITPR1 in patient 36. (F) Electropherograms depicting the mutations of DYNC1H1 and DCTN1 in patient 37 and her parents. Arrows indicate the mutated nucleotides. The left three panels indicating c.11824C>T (p.P3942S) of DYNC1H1 show that the mutation is inherited from the mother, and the right three panels indicating c.497C>G (p.S166C) of DCTN1 show the mutation is inherited from the father.