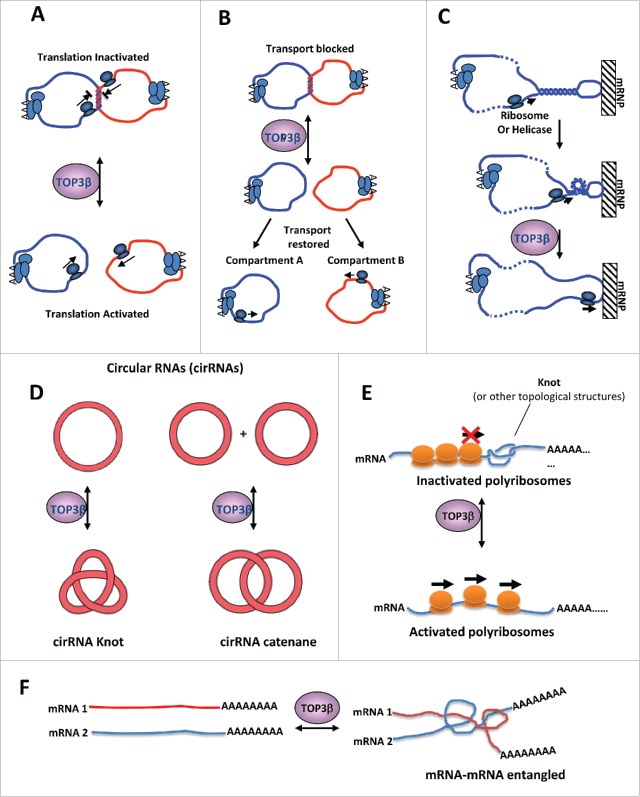
Figure 4.
Models for why RNA metabolism may create topological problems that need an RNA topoisomerase to solve. (A)(B) Models to illustrate that Top3β may catalyze catenation or decatenation of 2 mRNA circles, thus enhancing or inhibiting translation (B) or transport of mRNAs (C). It has been shown that mRNA circles can form when 2 ends of mRNA interacting with a common protein. (C) A model illustrates that helical torsions may arise when a translating ribosome or a helicase unwinds a duplex region in an mRNA hairpin. If the hairpin is bound to an immobile mRNP or cellular matrix, the helical torsion will not be relieved by rotation. Such torsion may impede progression of ribosomes or RNA helicases during translation or transport. Top3β can relax the helical torsion through its strand passage activity. (D) Top3β may interconvert circular RNAs to knots or catenane structures, which may affect the function of these RNAs. (E) A linear mRNA may form an intramocular knot, which may block progression of translating ribosomes. Top3β may catalyze knotting or unknotting of the linear mRNA, and thus affect translation. (F) Two linear mRNAs get entangled with each other, which may interfere with their functions. Top3β may catalyze entangling or segregation of these mRNAs, thus affecting their functions. This figure was adapted from Fig. S7 of a previous publication.17