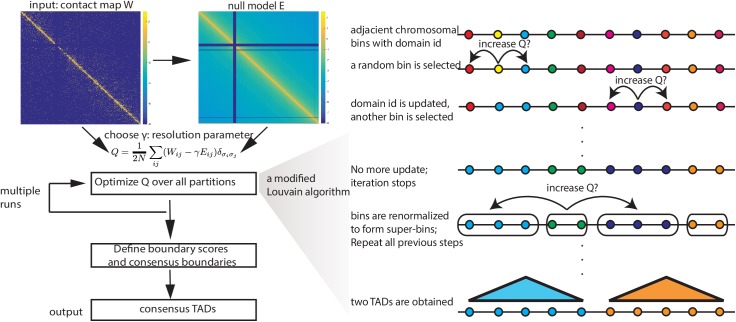
Fig 1. Overview of MrTADFinder.
The input of MrTADFinder is an intra-chromosomal contact map W. A null model E is obtained from W. Given a particular resolution γ; the chromosome is partitioned probabilistically in a way such that the objective function Q is maximized. The optimization is performed by a modification Louvain algorithm shown on the right. The algorithm is stochastic because the updating order of nodes is random. A boundary score is defined after multiple trials for all adjacent bins. Adjacent bins that are robustly assigned to two different TADs form a consensus boundary. The output of MrTADFinder is a set of consensus domains bound by the consensus domains.