Figure 2.
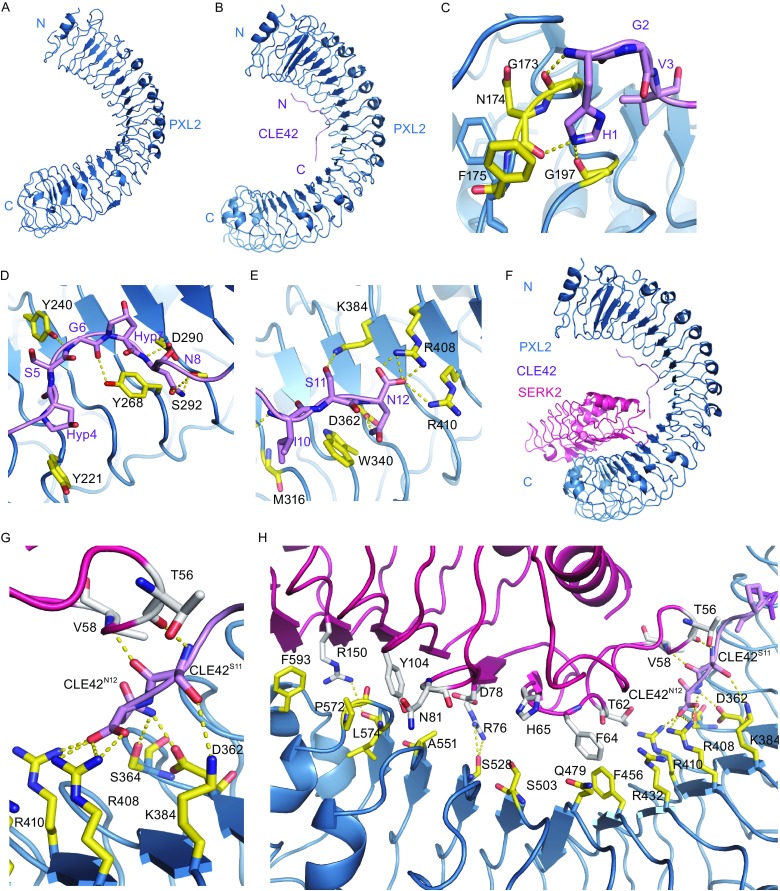
Modeled structure of CLE42-PXL2 LRR and PXL2LRR-CLE42-SERK2LRR. (A) Crystal structure of PXL2LRR alone shown in cartoon. (B) Modeled structure of PXL2LRR-CLE42 complex using as the PXYLRR-CLE41 template. CLE42 adopts an “Ω”-like kink and binds to the concave surface of PXL2LRR. (C) Detailed interaction of the N-terminal side of CLE42 with PXL2LRR. The side chains of some amino acids from CLE42 and PXL2LRR are shown in violet and yellow orange, respectively. Yellow dashed lines indicate hydrogen bonds or salt bridges. (D) Detailed interaction of the central region of CLE42 with PXL2LRR. (E) Detailed interaction of the C-terminal side of CLE42 with PXL2LRR. (F) Model structure of PXL2LRR-CLE42-SERK2LRR complex. (G) Detailed interaction between the C-terminal residues Ser11 and Asn12 of CLE42 and PXL2, SERK2. The side chains of some amino acids from CLE42, PXL2LRR, and SERK2LRR are shown in violet, yellow orange, and pink, respectively. Yellow dashed lines indicate hydrogen bonds or salt bridges. (H) Detailed interaction between PXL2LRR-CLE42 and SERK2LRR
