Abstract
Background
Familial eosinophilia (FE) is a rare autosomal dominant inherited disorder characterized by the presence of lifelong peripheral eosinophilia (>1500/μL). Mapped to chromosome 5q31-q33, the genetic cause of FE is unknown, and prior studies have failed to demonstrate a primary abnormality in the eosinophil lineage.
Objective
The aim of the present study was to identify the cells driving the eosinophilia in FE.
Methods
Microarray analysis and real-time PCR were used to examine transcriptional differences in peripheral blood mononuclear cells (PBMC), and in purified cell subsets from affected and unaffected family members belonging to a single large kindred. Cytokine levels in serum and PBMC culture supernatants were assessed by suspension array multiplexed immunoassays.
Results
Whereas IL5 mRNA expression was significantly increased in freshly isolated PBMC from affected family members, this was not accompanied by increased mRNA expression of other Th2 cytokines (IL4 or IL13). Serum levels of IL-5 and IL-5 receptor α, but not IgE, were similarly increased in affected family members. Of note, IL5 mRNA expression was significantly increased in purified CD3+ CD4+, CD14+, CD19+ and ILC2 cells from affected family members, as were IL-5 protein levels in supernatants from both stimulated PBMC and ILC2 cultures.
Conclusions
These data are consistent with the hypothesis that the eosinophilia in FE is secondary to dysregulation of IL-5 production in PBMC (and their component subsets).
Keywords: Eosinophil, hypereosinophilic syndrome, autosomal dominant, cytokine, interleukin 5
Introduction
Familial eosinophilia (FE) is a disorder characterized by the presence of sustained peripheral eosinophilia >1500/μL beginning at birth that has an autosomal dominant inheritance pattern (1,2). Despite prolonged eosinophilia in affected family members, clinical manifestations related to the eosinophilia are uncommon (2,3). This “benign” phenotype is likely related to a relative lack of eosinophil activation as evidenced by cellular morphology, surface activation markers and release of eosinophil granule proteins (3).
Although the gene responsible for FE has been mapped by genome wide linkage analysis in one large kindred to a region on chromosome 5q containing the cytokine gene cluster (multipoint LOD score of 6.49), targeted sequencing of candidate genes, including IL5, IL3, and GMCSF, and their promoters has failed to identify the causative mutation (4). Attempts to identify the causative mutation by whole genome sequencing are ongoing, but have been unsuccessful to date.
The increased levels of morphologically normal peripheral blood eosinophils observed in subjects with FE could be due to a primary abnormality in the eosinophil lineage or to mediators produced by cell types other than the eosinophil. Prior studies, including morphologic examination, ex vivo differentiation of eosinophils from CD34+ cells, and eosinophil survival assays, have provided little evidence for a primary eosinophil defect (3). Although serum cytokine levels measured previously were similar between affected and unaffected family members, the assay used to measure IL-5 was relatively insensitive, detecting IL-5 in sera from only 3/14 affected and 3/22 unaffected family members (1). The aim of the present study was to identify the cell population(s) responsible for driving the eosinophilia in FE.
Patient population and methods
Study subjects
Affected and unaffected family members from two kindreds with autosomal dominant hypereosinophilia (HE; absolute eosinophil count >1500/μL) documented over at least three generations were evaluated on institutional review board-approved clinical protocols to study familial eosinophilia (NCT00091871 and a prior protocol that preceded clinicaltrials.gov) (see pedigrees in Supplemental Figure 1). Healthy controls (HC) without eosinophilia were recruited on a separate institutional review board-approved clinical protocol to obtain normal blood samples for in vitro research (NCT00001846). All participants gave written informed consent. As described previously (3), subjects underwent an extensive clinical evaluation to exclude secondary causes of eosinophilia and assess end organ manifestations of eosinophilia. Routine laboratory testing, including complete blood counts, serum immunoglobulin levels and assessment of T, B and NK cell subsets in whole blood by flow cytometry, was performed in the Department of Laboratory Medicine, NIH Clinical Center. Mast cells were quantified in bone marrow core biopsies by a single hematopathologist following tryptase immunostaining. Results are reported as the average count over 10 high-powered fields.
PBMC purification
PBMC were isolated from peripheral blood by density gradient centrifugation (Ficoll-Paque PLUS, GE Healthcare, Uppsala, Sweden). Red cells were lysed for 10 minutes at room temperature with ammonium-chloride-potassium (ACK) lysing buffer (Invitrogen, Carlsbad, CA). Cell number and viability were determined by staining with trypan blue. Aliquots of PBMC were cryopreserved in liquid nitrogen in freezing medium with C–RPMI containing 10% fetal bovine serum (FBS; Invitrogen) and 7.5 % dimethyl sulfoxide (DMSO; Thermo Fisher Scientific) or stored in Trizol (Invitrogen) at -80°C for future use.
Magnetic enrichment of CD3+ and CD3- PBMC subsets
PBMC (either fresh or from previously cryopreserved samples) from affected and unaffected family members were magnetically labeled with CD3 Microbeads (Miltenyi Biotec). Using positive selection in an autoMACS Pro separator, the magnetically labeled CD3+ T cells and the unlabeled CD3- T cells were isolated.
Flow sorting of PBMC subsets
PBMC from affected family members and healthy controls were stained with antibodies to CD3, CD4, CD8, CD19 and CD14 for isolation of CD3+CD4+ and CD3+CD8+ T cells, CD19+ B cells and CD14+ monocytes. Fresh PBMC were stained with a cocktail of lineage markers to deplete lineage positive cells and with subset markers to sort innate lymphoid cells (ILC) subsets. ILCs were defined as LIN-CD45+CD127+ and ILC subsets as ILC1 (CD117- CRTH2-), ILC2 (CRTH2+) and ILC3 (CRTH2- CD117+) as previously described (5). A complete listing of antibodies used can be found in Supplemental Table 1.
All cells were sorted using a FACSAriaTM II cell sorter (BD Biosciences) according to the manufacturer's instructions. The gating strategies are provided in Supplementary Figure 2.
Microarray analysis
Total RNA was isolated from freshly purified PBMC stored in Trizol according to the manufacturer's instructions (Invitrogen) prior to reverse-transcription using a T7 oligo dT (24) primer and Superscript II (Invitrogen). Second-strand cDNA synthesis was obtained with RNase H, E. coli DNA polymerase I, and DNA ligase (Invitrogen). cDNA was blunt-ended with T4 DNA polymerase (Invitrogen) and purified using the QIAQuick PCR purification Kit™ (Qiagen, Germantown, MD). Labeling was performed using the ENZO BioArray™ HighYield™ RNA Transcript Kit (Affymetrix) according to the manufacturer's instructions. After purification, labeled cRNA was quantified by OD, and the quality was assessed on a Bioanalyzer (Agilent, Santa Clara, CA). Fragmented cRNA with hybridization controls and Oligo B2 from Affymetrix were hybridized on Affymetrix U133 Plus 2.0 Genechips for 18 hours followed by washing and staining on Affymetrix Fluidic 400. The arrays were scanned using the Affymetrix GeneChip Scanner 3000. Array images were analyzed using Affymetrix Gene Chip Operating System (GCOS) software using the MAS5 algorithm. The MAS5 algorithm (6) computes an adjusted signal level for each probe set to estimate expression of the target mRNA of that probe set. Values were normalized for each probe set across all arrays in the dataset.
Real-Time qRT-PCR analysis of PBMC and of PBMC subsets
Total RNA was extracted from PBMC or from flow sorted PBMC subsets in Trizol according to the manufacturer's instructions (Invitrogen). Total RNA (1 μg) was reverse-transcribed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). qRT-PCR was carried out using TaqmanR Fast Universal PCR Master Mix using commercially available FAM-MGB primer probes (TaqmanR Gene Expression Assay, Applied Biosystems) for IL5 (Hs00174200_m1), IL13 (Hs00174379_m1), IL4 (Hs00174122_m1) and IFN-γ (Hs00174143_m1). All reactions were performed in triplicate, and the relative expression was normalized to the corresponding 18S rRNA cycle threshold (Ct). Lack of expression was defined as a Ct of more than 35 cycles (for IL5) despite the presence of sufficient cDNA (18S Ct <12). Data are expressed as 1/ΔCt.
Measurement of soluble mediators in serum and PBMC supernatants
Serum was collected from whole blood by centrifugation and stored immediately at −80°C for future use. Isolated PBMC were thawed, washed and resuspended in RPMI 1640 medium, supplemented with gentamicin, L-glutamine (2mM), HEPES (10mM), sodium pyruvate (all from Invitrogen, SanDiego, CA) in 24-well tissue culture plates (Costar, Corning, NY). After resting overnight, the cells were cultured with and without PMA and ionomycin (final concentrations: 15 ng/mL and 150 ng/ml, respectively) for 18 hours at 37°C. The culture supernatants were harvested and stored at -80 °C.
Flow-sorted ILC2 cells were cultured in 96-well round bottom plates at a concentration of 2×103/150 μL/well in X-Vivo™ 15 medium (Lonza) supplemented with 1% heat-inactivated human AB serum, 10 U/mL of IL-2 (PeproTech) and 50 ng/mL of rIL-7 (PeproTech) and activated with PMA and ionomycin for 18 hours. Supernatants were collected at day 2 and stored at -80°C.
Levels of IL-4, IL-5, IL-13, GM-CSF, and eotaxin1/CCL11 were measured in both serum and supernatants by suspension array multiplex immunoassays (Millipore). All assays were run in duplicate. The lower limits of detection were: IL-5 (0.1 pg/ml), IL-13 (0.4 pg/ml), IL-4 (0.6 pg/ml), IFN-γ (0.1pg/mL), GM-CSF (9.5 pg/ml) and eotaxin1/CCL11 (1.2 pg/ml). Serum TARC levels were measured by ELISA (Human CCL17/TARC DuoSet ELISA, R&D Systems) according to the manufacturer's instructions. Soluble IL-5 receptor alpha (sIL-5Rα) levels were quantified in serum using an in-house chemiluminescence capture ELISA as previously described (7).
Statistical analysis
Geometric means (GM) were used for measurements of central tendency. Group means were compared using the Mann-Whitney test, and paired samples were analyzed using the Wilcoxon signed rank test. A p-value of <0.05 was considered statistically significant for all tests. For the microarray data, the table of gene probe set expression estimates (using the MAS5 algorithm) for 17 arrays (9 affected subjects, 8 unaffected) was transformed by quantile normalization before calculating expression differences between the two groups using the SAS mixed-effects ANOVA procedure in JMP/Genomics version 8.0 (SAS Institute, Cary NC). P-values were adjusted for multiple testing using the False Discovery Rate (FDR) method (8).
Results
Increased IL5 mRNA expression in PBMC from affected family members
To investigate the role of PBMC in driving the eosinophilia in FE, gene expression microarray analysis was performed using PBMC RNA from 9 affected and 8 unaffected members of the previously described family with autosomal dominant FE mapped to chromosome 5q31-33 (Klion Blood 2004). Restricting the analysis to the probe sets for the 313 genes in the previously mapped region, the highest differential expression (after a p=0.05 positive or negative fold-change of 2 cutoff) between affected and unaffected family members was seen for the following genes: IL5, ARHGEF37, CSF2, ECSCR, FGF1, GRIA1, IL12B, IL4, MYOZ3, MZB1, NRG2, PCDHGC4, PDGFRB, PSD2, SPARC (Figure 1).
Figure 1.
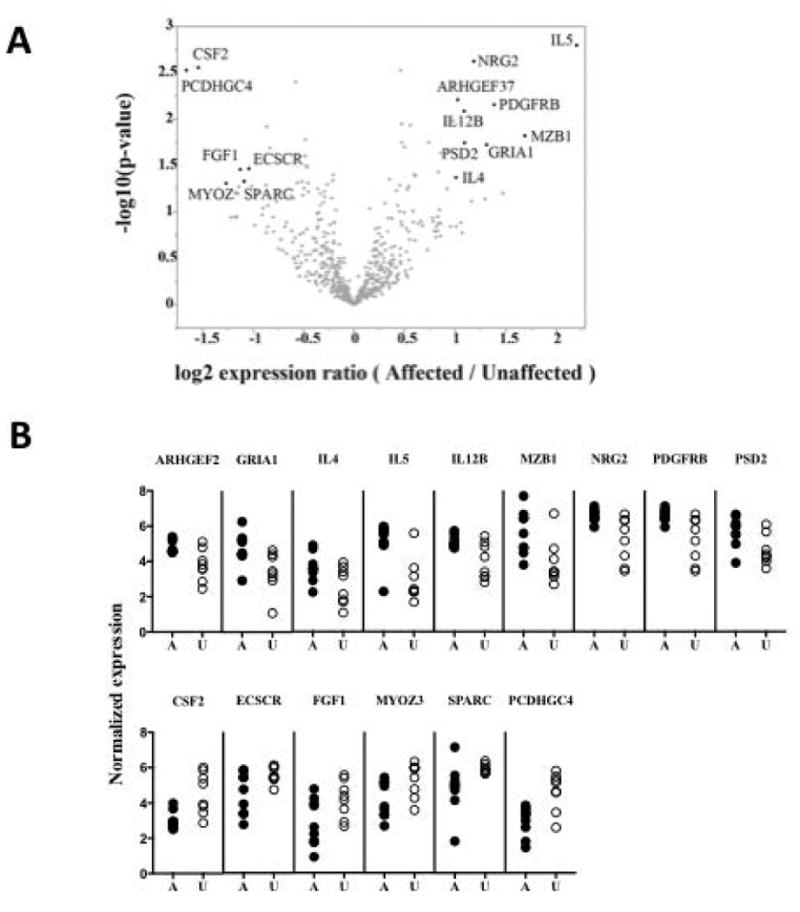
Gene expression analysis for chromosomal region [5q31.1 - 5q33.1]. (A) Results of ANOVA test for difference in geometric average expression. Probe sets with expression differences greater than 2–fold and (unadjusted) p-value < 0.05 are labeled with their corresponding gene symbol. The False Discovery Rate values for these selected genes ranged from 0.32 to 0.70. Thus, none of the expression differences in the region were judged be statistically significant once this correction for multiple testing was applied. (B) Individual subject expression levels for the selected, most differentially expressed, genes.
Given the dramatic increase in eosinophilia in FE and the known role of IL-5 in eosinophilia, IL5 mRNA expression in unstimulated PBMC was assessed next by qRT-PCR. As can be seen in Figure 2, constitutive expression of IL5 mRNA was detected in all 10 affected family members tested, in 8 of 9 unaffected family members and in 5 of 7 healthy controls (Figure 2A). Geometric mean IL5 mRNA expression was significantly increased in affected family members (1/ΔCt of 0.07 in affected family members vs. 0.03 in unaffected family members, and 0.04 in healthy controls, p<0.0001 and p<0.005, respectively). In contrast, mRNA from two other Th2 cytokines, IL13 and IL4 (also increased in affected family members based on the microarray analysis), was not detectable in any of the 8 affected or 8 unaffected family members tested using qRT-PCR. mRNA expression of the Th1 cytokine, IFNγ, was detectable in 10 affected and 8 unaffected family members, but was comparable between the two groups (1/ΔCt of 0.05 in affected and unaffected family members) (Figure 2B). mRNA expression of RAD50, a gene in close proximity to IL5, was also comparable between the two groups (Supplemental Figure 3).
Figure 2.
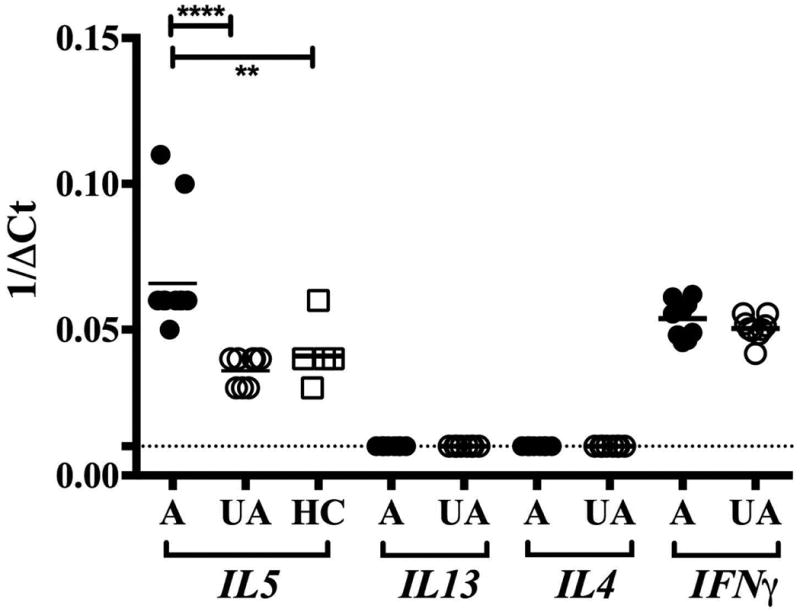
Cytokine mRNA expression by PBMC in FE. IL5, IL4, IL13 and IFNγ mRNA expression by unstimulated PBMC as assessed by qRT-PCR is shown as 1/ΔCt for affected (A) and unaffected (UA) family members and healthy controls (HC). Horizontal lines represent the geometric means for each group. **p<0.005 and ****p<0. 0001 as compared to affected family members.
Increased serum levels of IL-5 and sIL5-Rα in affected family members
Similar to the mRNA expression data, serum IL-5 levels were low, but detectable, in all 13 of the affected family members tested and in 10/14 of the unaffected family members (p=NS, Fisher's exact test). However, geometric mean serum IL-5 levels were significantly increased in affected family members (GM 4.45 pg/mL vs. 1.60 pg/mL in unaffected family members; p<0.05). Serum levels of sIL-5Rα, a surrogate marker of cytokine-driven eosinophilia (7), were also significantly increased in affected family members (GM 450.9 ng/mL vs. 174.9 ng/mL in unaffected family members; p<0.01). Serum levels of GM-CSF, another cytokine that plays a role in the induction of eosinophilia and has been shown to increase serum levels of sIL-5Rα, was significantly decreased in affected family members (GM 0.37 pg/mL vs. 1.33 pg/mL in unaffected family members; p=0.02). Surprisingly, serum levels of the eosinophil-attracting chemokine, eotaxin1/CCL11, were also decreased in affected family members (GM 128.6 pg/mL vs. 241.2 pg/mL in unaffected family members, p=0.02).
To explore whether the increased IL-5 was part of a more generalized Type 2 response, serum levels of the type 2 cytokine, IL-13 and IL-4, as well as serum IgE levels were assessed. Serum levels of IL-13 and IgE were significantly decreased in affected family members (GM serum IL-13 level of 1.52 pg/mL in affected vs. 5.02 pg/mL in unaffected family members and GM serum IgE level of 19.41 IU/mL in affected vs. 57.23 IU/mL in unaffected family members; p=0.01 and p=0.03, respectively; Figure 3). Serum IL-4 levels were not measurable in serum from any of the subjects tested. Serum levels of TARC/CCL17, a marker of T cell activation, and the Th1 cytokine, IFN-γ, were similar between the two groups (Figure 3).
Figure 3.
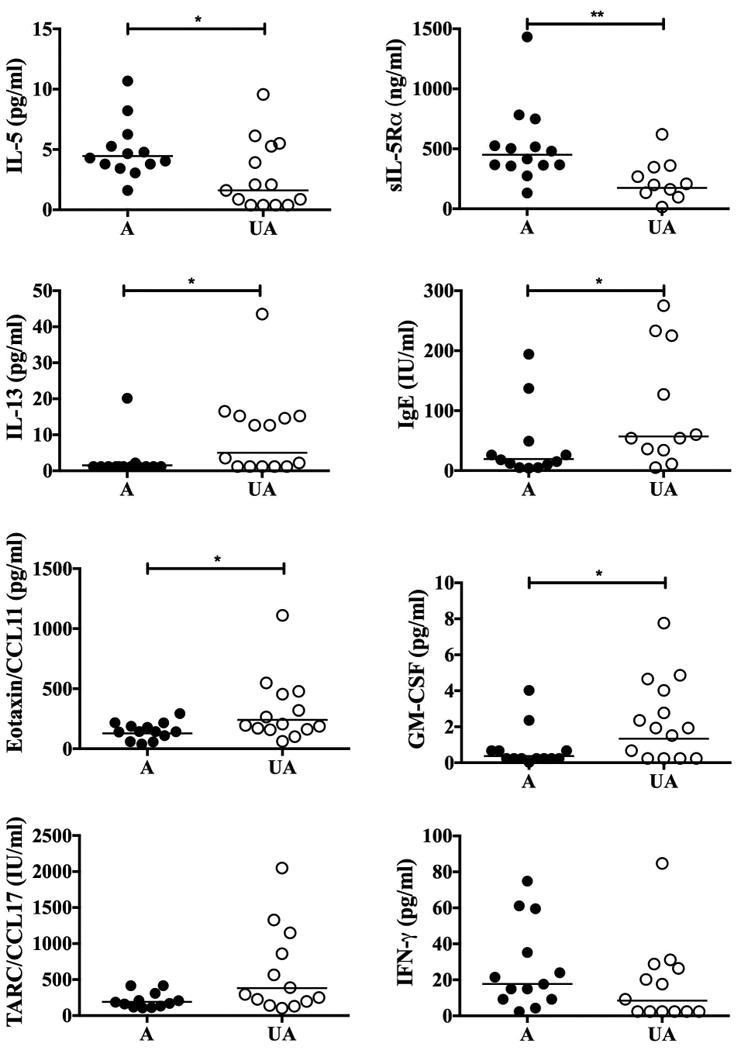
Serum mediator levels in FE. Serum levels of IL-5, sIL-5Rα, IL-13, IgE, eotaxin1/CCL11, GM-CSF, TARC/CCL17 and IFN-γ are shown for individual affected (A) and unaffected (UA) family members. The horizontal lines represent the geometric means for each group. *p<0.05, **p<0.005
Increased release of IL-5 following PMA/ionomycin stimulation of PBMC from affected family members
Although IL-5 levels tended to be higher in the supernatants from PMA/ionomycin-stimulated PBMC from affected family members, the difference did not reach statistical significance (GM 221.4 pg/mL vs. 26.49 pg/mL in unaffected family members; p=0.059). Stimulated PBMC supernatant levels of IL-4 (GM 73.46 pg/mL in affected vs. 191.4 pg/mL in unaffected family members) and IL-13 (GM 186.5 pg/mL in affected and 302.5 pg/mL in unaffected) were comparable between the two groups (Figure 4). No IL-5, IL-4 or IL-13 was detectable in supernatants from unstimulated PBMC.
Figure 4.
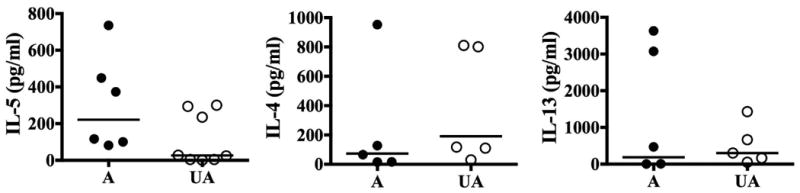
Cytokine levels in supernatants from mitogen-stimulated PBMC in FE. Levels of IL-4, IL-5, and IL-13 in PMA/ionomycin stimulated PBMC supernatants are shown for individual affected (A) and unaffected (UA) family members. The horizontal lines represent the geometric means for each group.
Increased expression of IL5 mRNA is not restricted to CD4+ T cells in affected family members
Since CD4+ T cells are known to be a major source of IL-5, the percentages and absolute numbers of CD3+, CD4+, CD8+, CD4+CD3+, CD8+CD3+, CD3-CD4- T cells, CD3+HLADR+, CD3+CD25+, CD20+, CD3-CD16+CD56+ NK cells and CD3+CD16+CD56+ NKT cells were assessed by whole blood flow cytometry to determine whether alterations of particular PBMC components/subsets might be the reason for the observed increased IL5 expression. No significant differences were detected between the affected and unaffected family members (Supplementary Figure 4).
As a next step, cDNA was prepared from magnetic bead-enriched CD3+ and CD3- cells and from flow-sorted CD3+CD4+, CD3+CD8+, CD3-CD14+ and CD3-CD19+ subsets from affected and unaffected family members. IL5 mRNA was significantly upregulated in both CD3+ and CD3- subsets from affected family members (Figure 5A, p<0.01). Furthermore IL5 mRNA was detectable, albeit at low levels, in flow-sorted CD4+, CD8+, CD14+ and CD19+ from all 6 affected family members and none of the 3 unaffected family members tested (Figure 5B, p<0.05 for all subsets).
Figure 5.
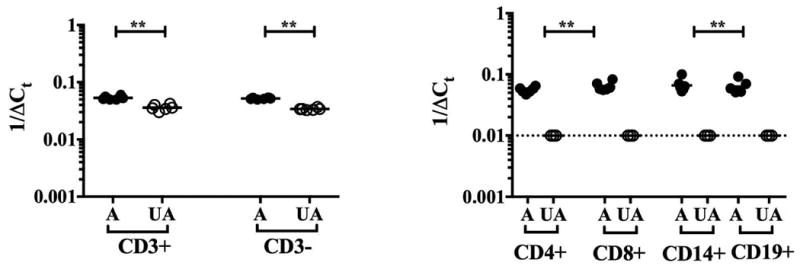
IL5 mRNA expression by PBMC subsets in FE. IL5 mRNA expression by unstimulated PBMC subsets as assessed by qRT-PCR is shown as 1/ΔCt for individual affected (A) and unaffected (UA) family members. The horizontal lines represent the geometric means for each group. *p<0.05 as compared to affected family members.
Since ILCs are a known source of IL-5 and could account for the increased IL5 seen in the CD3- PBMC fraction, ILC subsets were assessed in affected family members (n=3) and compared to those from normal controls (n=6). Whereas the ILC2 subset accounted for a similar proportion of the total ILC population in the two groups (GM 7% (range 2-10%) in healthy controls compared to 4% (range 2-10%) in the affected family members; Figure 6A), flow-sorted ILC2s from the affected family members produced increased amounts of IL-5 following stimulation with PMA/ionomycin (GM net IL-5 of 185.9 vs. 6.194 in healthy controls; Figure 6B).
Figure 6.
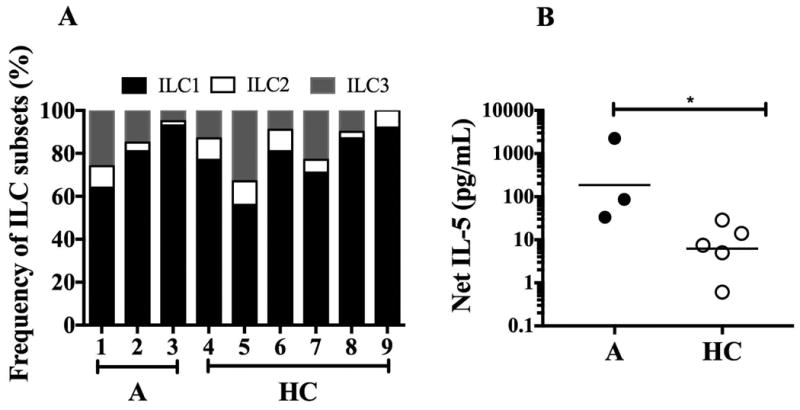
ILC subset frequency and net IL-5 production in FE. A) Percentage distribution of ILC1, ILC2 and ILC3 subsets are shown for individual healthy controls (HC) and affected (A) family members. B) Net IL-5 release from unstimulated and PMA/Ionomycin stimulated, flow-sorted ILC2 subsets. Horizontal lines represent the geometric means for each group. *p<0.05 as compared to affected family members.
Discussion
Familial eosinophilia (FE) is a rare genetic disorder in which seemingly normal eosinophils are increased in the peripheral blood without clinical consequences. Although the gene has been mapped to chromosome 5q31-q33 (4), the causative mutation has not been identified. Whereas absolute eosinophil counts (AEC) have been linked to chromosome 5q31-q33 in other settings, including asthma in children (9,10), schistosomiasis (11) and, more recently, in adults with a high risk of cardiovascular disease (12), little is known about the specific mechanisms underlying these associations. Furthermore, the complex genetics of these conditions and the presence of elevated IgE and other findings suggestive of a more generalized Type 2 response, complicate the interpretation of functional studies. In this context, FE provides a unique opportunity to study the effect of a single genetic abnormality in chromosome 5q31-q33 on the regulation of eosinophilia. The major limitation of the present study is the fact that the studies to date have been restricted to one large kindred. Although a second family with asymptomatic eosinophilia over 3 generations and increased production of IL-5 has recently been identified, the small number of affected family members available for study and logistical constraints have precluded additional analyses.
Given the lack of evidence pointing to an abnormality in the eosinophil lineage in FE, the current study focused on the role of circulating mononuclear cells in driving the eosinophilia. Although microarray analysis revealed few differences in PBMC gene expression between affected and unaffected family members, a relative increase in IL5 expression was observed and confirmed by qRT-PCR. Of note, mild but significant increases in basophils and mast cells, the only other lineages that express IL-5Rα in humans, were also seen in affected family members. Other lineages were similar between the two groups (Supplemental Figure 5). Increased IL5 expression was also significant at the protein level in serum and in supernatants from stimulated PBMCs. In contrast to the generalized Th2 responses seen in allergen or helminth-driven eosinophilia, however, there was no concomitant increase in PBMC expression of IL4 or IL13 mRNA, and serum IL-13 and IgE levels were significantly decreased in affected family members.
The genes for the three major Th2 cytokines, IL5, IL4 and IL13, are clustered in a 160 kb region of chromosome 5 and are most often expressed together (13,14). Nevertheless, selective regulators of expression of IL4 and IL13 have been described (15). Moreover, Th2 cells producing IL-4 and IL-13, but not IL-5, have been demonstrated by flow cytometry in normal and atopic individuals (16) as well as in children with measles (17) and adult patients with filarial infection (18). In contrast, prior to the present study, selective production of IL-5 had only been convincingly demonstrated in murine T cell clones stimulated with IL-2 (19) and in clonal T cell populations from some patients with lymphocytic variant HES (20,21).
Whereas coordinated expression of IL4, IL5 and IL13 has been shown to be regulated by a locus control region (LCR) containing the 3′ portion of the RAD50 gene (14,22), the genes encoding IL4 and IL13 are adjacent to each other and to the LCR, but separated from the IL5 gene by the single large gene encoding RAD50. Furthermore, IL5 transcription occurs in a direction opposite from that of IL4 and IL13. Consequently, it is not difficult to imagine a scenario in which specific regulatory elements might lead to isolated expression of IL5 (23). In fact, isolated upregulated IL5 transcription has been shown to occur in response to mIR-1248 (24). In the present study, the lack of concomitant upregulation of RAD50 is consistent with the hypothesis that IL5 is not being expressed under control of the previously described LCR and suggests a novel regulatory mechanism for IL5 mRNA expression. The fact that all cell types examined expressed increased levels of IL5 mRNA implies that this dysregulation of expression is lineage-independent.
In summary, FE is a rare genetic disorder of unknown etiology characterized by persistent eosinophilia and increased IL-5 production by peripheral blood mononuclear cells. The central role of IL-5 in the production, activation and survival of eosinophils has made it a prime target for therapeutic intervention in asthma and other eosinophilic disorders. FE provides a unique opportunity to explore a novel mechanism of IL-5 regulation in humans. Ultimately, this could lead to the development of new therapeutic strategies for the treatment of eosinophilic disorders.
Supplementary Material
Acknowledgments
The authors would like to thank the many members of the Clinical Parasitology Section of the Laboratory of Parasitic Diseases who have participated in the protocol management and patient care that made this study possible.
Funding: The work was funded by the Division of Intramural Research, NIAID, NIH. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No.HHSN261200800001E.
Abbreviations
- A
Affected family members
- Ct
Cycle threshold
- FE
Familial eosinophilia
- FHES
Familial hypereosinophilic syndrome
- GM
Geometric mean
- HD
Healthy controls
- HE
Hypereosinophilia
- LOD
Logarithm of Odds
- PBMC
Peripheral blood mononuclear cells
- qRT-PCR
Quantitative reverse transcriptase polymerase chain reaction
- UA
Unaffected family members
Footnotes
Author Contributions: Senbagavalli Prakash Babu, Yun-Yun K. Chen, Sandra Bonne-Annee, Jun Yang, and Timothy Myers participated in the design and execution of the experiments, analysis of the data and the writing of the paper, Thomas Nutman and Amy Klion participated in the design and analysis of the experiments and the writing and editing of the paper.
Conflict of Interest: None of the authors have a declared conflict of interest.
References
- 1.Lin AY, Nutman TB, Kaslow D, Mulvihill JJ, Fontaine L, White BJ, et al. Familial eosinophilia: clinical and laboratory results on a U.S. kindred. Am J Med Genet. 1998 Mar 19;76(3):229–37. [PubMed] [Google Scholar]
- 2.Naiman JL, Oski FA, Allen FH, Diamond LK. Hereditary eosinophilia: report of a family and review of the literature. Am J Hum Genet. 1964 Jun;16(2):195–203. [PMC free article] [PubMed] [Google Scholar]
- 3.Klion AD, Law MA, Riemenschneider W, McMaster ML, Brown MR, Horne M, et al. Familial eosinophilia: a benign disorder? Blood. 2004 Jun 1;103(11):4050–5. doi: 10.1182/blood-2003-11-3850. [DOI] [PubMed] [Google Scholar]
- 4.Rioux JD, Stone VA, Daly MJ, Cargill M, Green T, Nguyen H, et al. Familial eosinophilia maps to the cytokine gene cluster on human chromosomal region 5q31-q33. Am J Hum Genet. 1998 Oct;63(4):1086–94. doi: 10.1086/302053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mjösberg JM, Trifari S, Crellin NK, Peters CP, van Drunen CM, Piet B, et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol. 2011 Nov;12(11):1055–62. doi: 10.1038/ni.2104. [DOI] [PubMed] [Google Scholar]
- 6.Pepper SD, Saunders EK, Edwards LE, Wilson CL, Miller CJ. The utility of MAS5 expression summary and detection call algorithms. BMC Bioinformatics. 2007 Jul 30;8:273. doi: 10.1186/1471-2105-8-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson TM, Maric I, Shukla J, Brown M, Santos C, Simakova O, et al. IL-5 receptor α levels in patients with marked eosinophilia or mastocytosis. J Allergy Clin Immunol. 2011 Nov;128(5):1086–92.e1. doi: 10.1016/j.jaci.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benjamini Y, Hochberg Y. On the Adaptive Control of the False Discovery Rate in Multiple Testing With Independent Statistics. Journal of Educational and Behavioral Statistics. 2000 Jan 1;25(1):60–83. [Google Scholar]
- 9.Palmer LJ, Daniels SE, Rye PJ, Gibson NA, Tay GK, Cookson WO, et al. Linkage of chromosome 5q and 11q gene markers to asthma-associated quantitative traits in Australian children. Am J Respir Crit Care Med. 1998 Dec;158(6):1825–30. doi: 10.1164/ajrccm.158.6.9804037. [DOI] [PubMed] [Google Scholar]
- 10.Martinez FD, Solomon S, Holberg CJ, Graves PE, Baldini M, Erickson RP. Linkage of circulating eosinophils to markers on chromosome 5q. Am J Respir Crit Care Med. 1998 Dec;158(6):1739–44. doi: 10.1164/ajrccm.158.6.9712040. [DOI] [PubMed] [Google Scholar]
- 11.Rodrigues V, Abel L, Piper K, Dessein AJ. Segregation analysis indicates a major gene in the control of interleukine-5 production in humans infected with Schistosoma mansoni. Am J Hum Genet. 1996 Aug;59(2):453–61. [PMC free article] [PubMed] [Google Scholar]
- 12.McLeod O, Silveira A, Valdes-Marquez E, Björkbacka H, Almgren P, Gertow K, et al. Genetic loci on chromosome 5 are associated with circulating levels of interleukin-5 and eosinophil count in a European population with high risk for cardiovascular disease. Cytokine. 2016 May;81:1–9. doi: 10.1016/j.cyto.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly BL, Locksley RM. Coordinate regulation of the IL-4, IL-13, and IL-5 cytokine cluster in Th2 clones revealed by allelic expression patterns. J Immunol. 2000 Sep 15;165(6):2982–6. doi: 10.4049/jimmunol.165.6.2982. [DOI] [PubMed] [Google Scholar]
- 14.Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003 Jul;19(1):145–53. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka S, Motomura Y, Suzuki Y, Yagi R, Inoue H, Miyatake S, et al. The enhancer HS2 critically regulates GATA-3-mediated Il4 transcription in T(H)2 cells. Nat Immunol. 2011 Jan;12(1):77–85. doi: 10.1038/ni.1966. [DOI] [PubMed] [Google Scholar]
- 16.Upadhyaya B, Yin Y, Hill BJ, Douek DC, Prussin C. Hierarchical IL-5 expression defines a subpopulation of highly differentiated human Th2 cells. J Immunol. 2011 Sep 15;187(6):3111–20. doi: 10.4049/jimmunol.1101283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moss WJ, Ryon JJ, Monze M, Griffin DE. Differential regulation of interleukin (IL)-4, IL-5, and IL-10 during measles in Zambian children. J Infect Dis. 2002 Oct 1;186(7):879–87. doi: 10.1086/344230. [DOI] [PubMed] [Google Scholar]
- 18.Anuradha R, George PJ, Hanna LE, Chandrasekaran V, Kumaran PP, Nutman TB, et al. Parasite-antigen driven expansion of IL-5(-) and IL-5(+) Th2 human subpopulations in lymphatic filariasis and their differential dependence on IL-10 and TGFβ. PLoS Negl Trop Dis. 2014 Jan 30;8(1):e2658. doi: 10.1371/journal.pntd.0002658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohjanen PR, Okajima M, Hodes RJ. Differential regulation of interleukin 4 and interleukin 5 gene expression: a comparison of T-cell gene induction by anti-CD3 antibody or by exogenous lymphokines. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5283–7. doi: 10.1073/pnas.87.14.5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roufosse F, Schandené L, Sibille C, Willard-Gallo K, Kennes B, Efira A, et al. Clonal Th2 lymphocytes in patients with the idiopathic hypereosinophilic syndrome. Br J Haematol. 2000 Jun;109(3):540–8. doi: 10.1046/j.1365-2141.2000.02097.x. [DOI] [PubMed] [Google Scholar]
- 21.Simon HU, Plötz SG, Dummer R, Blaser K. Abnormal clones of T cells producing interleukin-5 in idiopathic eosinophilia. N Engl J Med. 1999 Oct 7;341(15):1112–20. doi: 10.1056/NEJM199910073411503. [DOI] [PubMed] [Google Scholar]
- 22.Koh BH, Hwang SS, Kim JY, Lee W, Kang M-J, Lee CG, et al. Th2 LCR is essential for regulation of Th2 cytokine genes and for pathogenesis of allergic asthma. Proc Natl Acad Sci U S A. 2010 Jun 8;107(23):10614–9. doi: 10.1073/pnas.1005383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Stry M, Bix M. Explaining discordant coordination. Nat Immunol. 2011 Jan;12(1):16–7. doi: 10.1038/ni0111-16. [DOI] [PubMed] [Google Scholar]
- 24.Panganiban RPL, Pinkerton MH, Maru SY, Jefferson SJ, Roff AN, Ishmael FT. Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. Am J Clin Exp Immunol. 2012 Nov 15;1(2):154–65. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


