Abstract
Monoclonal antibody technologies have enabled dramatic advances in immunology, the study of infectious disease, and modern medicine over the past 40 years. However, many monoclonal antibody discovery procedures are labor- and time-intensive, low efficiency, and expensive. Here we describe an optimized mAb discovery platform for the rapid and efficient isolation, cloning and characterization of monoclonal antibodies in murine systems. In this platform, antigen-binding splenic B cells from immunized mice are isolated by FACS and cocultured with CD40L positive cells to induce proliferation and mAb production. After 12 days of coculture, cell culture supernatants are screened for antigen, and IgG positivity and RNA is isolated for reverse-transcription. Positive-well cDNA is then amplified by PCR and the resulting amplicons can be cloned into ligation-independent expression vectors, which are then used directly to transfect HEK293 cells for recombinant antibody production. After 4 days of growth, conditioned medium can be screened using biolayer interferometry for antigen binding and affinity measurements. Using this method, we were able to isolate six unique, functional monoclonal antibodies against an antigen of the human malaria parasite Plasmodium falciparum. Importantly, this method incorporates several important advances that circumvent the need for single-cell PCR, restriction cloning, and large scale protein production, and can be applied to a wide array of protein antigens.
Keywords: B cell, antibody, monoclonal, recombinant, RACE, FACS, cloning
1. Introduction
The ability to efficiently isolate antigen-specific monoclonal antibodies (mAbs) has caused explosive growth in our understanding of antibody-mediated immunity, and has enabled the rapid generation of novel therapeutics and diagnostics (1). These advances were enabled by new methods to isolate natively matched heavy chain (HC) and light chain (LC) pairs from a single B cell, and the ability to reconstruct those native antibody sequences as recombinant mAbs in large scale production. To this end, monoclonal antibody development has matured rapidly over the last few years. Hybridoma technology, first developed over 40 years ago, is one widely used method for generating mAbs (2) that fuses antibody-producing primary B cells with myeloma cell lines. Through a laborious subcloning process, antibody-producing hybrids can be grown and screened for antigen binding (3, 4). One advantage of this method is that once isolated, a B cell hybridoma culture can be used to produce the desired antibody directly and the culture can be cryopreserved for indefinite future use. However, because the input B cells are typically not selected for antigen positivity, this method can require extensive screening to identify antigen-specific hybridomas (although recent advances in robotic screening have alleviated this somewhat) (5). In addition, the initial cell fusion efficiency can also be very low, despite recent refinements in methodologies (6).
Another method to generate mAbs is phage display, which randomly pairs HC and LC sequences to create a library of potential antibody combinations (1, 7). Libraries could be iteratively panned against the antigen of interest to select for antigen binding HC+LC combinations, but the native pairing of HC and LC from a native B cell is lost. Additionally, because the resultant antibodies are synthetic, there is increased risk of isolating self-reactive mAbs. Such methods were useful for generating reagents, but had limited utility to advance our understanding of humoral immunity in vivo. Additionally, such methods are extremely labor intensive and inefficient. Therefore, while useful in generating reagents and potential therapeutics, this method is not ideal for efforts to understand humoral immunity against vaccines or pathogens.
The development of methods to isolate and rescue natively matched heavy and light chain pairs from single antigen-positive B cells rapidly advanced the field (8–14). One key aspect of this strategy is that the initial step selects for antigen-positive B cells by fluorescence-activated cell sorting (FACS). Then, the heavy- and light-chain sequences are rescued by RT-PCR. In some methods, the sorted B cells are grown in culture and screened for binding prior to the rescue (8, 15, 16), although methods to do this with murine B cells are not well described. In other methods, the B cells are sorted directly into lysis buffer and the heavy- and light-chain sequences are amplified and sequenced without secondary screening (10–14, 17, 18). In such cases, the secondary screening takes place following the expression and production of recombinant mAbs. Importantly, single B cell isolation by FACS is translatable across species, and has been described for humans, rhesus macaques, rabbits, and mice (12, 19–22).
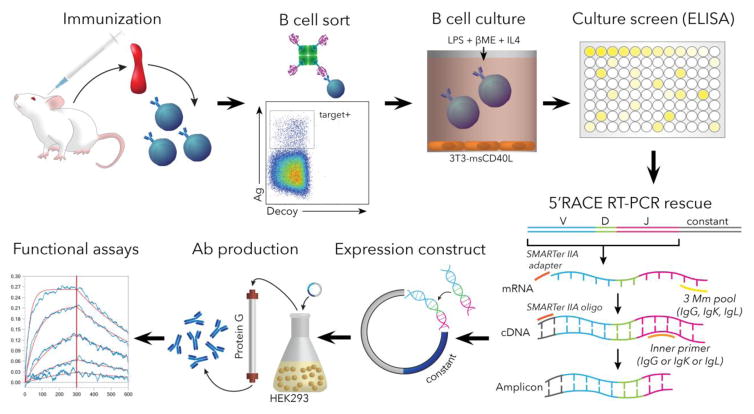
Here we report on a method for mAb cloning from single antigen-specific murine B cells (summarized in Fig. 1) as part of our ongoing efforts to evaluate vaccine candidates against pre-erythrocytic stages of malaria parasites. Our method utilizes FACS to sort antigen-specific B cells into cell culture under conditions that stimulate expansion and antibody production, where antibody binding can be screened directly from culture supernatants to ensure the preferential rescue of antigen-binding B cells. The BCR sequences are rescued from antigen binding wells by 5′RACE RT-PCR, cloned directly into antibody expression vectors, and expressed in small scale mammalian cell culture to verify antigen binding and perform affinity measurements. Several improvements in this protocol, including murine B cell co-culture, screening, one step cloning, expression, and small scale affinity measurements combine to create a straightforward protocol that ensures a high degree of success in mAb discovery. We have successfully deployed this method for cloning mAbs against multiple recombinant Plasmodium targets with similar efficiency. In an illustrative example, we show here that we used this method to successfully rescue mAbs that recognize a recombinant malaria parasite antigen with nanomolar affinities. Our findings show that this modular protocol will be useful for the rapid generation of mAbs and should be effective for a variety of target antigens and multiple per-target specificities.
Figure 1.
Workflow summary of mAb discovery pipeline in mice. Upon completion of the immunization regimen, spleens are harvested and gently homogenized to a single-cell suspension. The cell suspension is treated by negative magnetically-assisted selection until predominantly B cells remain. The B-cell suspension is stained using a cocktail of antibodies and Ag-streptavidin (as well as decoy-streptavidin) complexes to facilitate the sorting of Ag-positive class-switched cells. The cells are then cultured and induced to express antibodies in presence of supportive cytokines and feeder cells. Culture-conditioned supernatants containing target antibodies are then screened using Ag-specific ELISA, and positive wells are harvested for RNA isolation. Ab-encoding RNA is then reverse-transcribed, amplified and cloned into an expression cassette enabling recombinant production of the mAbs, followed by functional validation.
2. Materials and Methods
2.1 Production of recombinant antigen and fluorescent staining reagents
Plasmodium falciparum Thrombospondin-Related Anonymous Protein (PlasmoDB:PF3D7_1335900) (PfTRAP) was produced in suspension HEK293 cells cultures, as previously described (23, 24). The final expression construct contained a tPA signal (25), the extracellular domain of PfTRAP (amino acids 24–511), and a C-terminal HIS8 tag, followed by an AviTag (26). The expression constructs were cloned into pcDNA-3.4 vector (Thermo Fisher, Waltham, MA, USA) which uses a CMV immediate early promoter to drive transcription. Briefly, plasmid DNA encoding PfTRAP was introduced into HEK293 cells by high-density transfection with PEI MAX 40000 (Polysciences, Inc., Warrington, PA, USA)(27, 28). The cultures were grown in FreeStyle 293 serum-free media (Thermo Fisher) for five days before harvest. Upon harvest, the cells were removed from the cell supernatant by centrifugation, sodium azide was added at a final concentration of 0.02%, and NaCl was added to increase the concentration by 350 mM (we assumed that the final NaCl concentration was ~500 mM).
The protein was purified by a two-step chromatography protocol. The protein was captured using Ni-NTA (Qiagen, Germantown, MD, USA), washed in Wash buffer (25 mM Tris, pH 7.4, 300 mM NaCl, 20 mM imidazole) and eluted in Elution buffer (Wash buffer with 200 mM imidazole final concentration). The elution fractions were pooled, concentrated and separated by gel filtration on a standardized Superdex 200 16/600 (GE Healthcare, Chicago, IL, USA) column running in buffer HBS-E (10 mM Hepes, pH 7, 150 mM NaCl, 2 mM EDTA). Peak fractions were pooled, concentrated and stored at 4°C until use.
Biotinylation of AviTag-containing protein was carried out using a BirA biotin-protein ligase kit (Avidity, LLC; Aurora, CO, USA), according to manufacturer’s instructions. Following the biotinylation step, protein was purified by gel-filtration as above to remove free biotin, along with other reaction components, and concentrated. Complexation with streptavidin-conjugated fluorophores was performed by gradual mixing of 4 molar-equivalents of biotinylated PfTRAP with a 1 molar-equivalent streptavidin-fluorophore. The fluorescent biotinylated PfTRAP complex was created with streptavidin-BV785. In addition, a decoy staining reagent consisting of biotinylated P. yoelii circumsporozoite protein (PlasmoDB: PYYP_0405600) extracellular domain (amino acids 21–362) (PyCSP) expressed as described above (29, 30), and was used to generate a decoy staining reagent with streptavidin-BV510 (BioLegend, San Diego CA, USA).
2.2 Immunization of Balb/cj mice
To generate antigen-specific B cells, we immunized Balb/cj mice (obtained from Jackson Laboratories, Bar Harbor, ME, USA) with PfTRAP protein. Animals received three intramuscular injections spaced two weeks apart containing 25 μg of protein with 20% Adjuplex adjuvant (Invivogen, San Diego, CA, USA), followed by a final IV boost without adjuvant. 48–72 hours post final immunization, spleens were harvested and the splenocytes were processed for B cell sorting. All immunizations were carried out under protocols reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the Center for Infectious Disease Research (protocol NS02).
After harvesting, spleens were placed in a dish containing warm RPMI (Corning, Corning, NY) and dissociated using a 70-μm sieve (Falcon/Corning, Corning, NY, USA). The splenocytes were filtered through 70 μm Nitex nylon (Sefar, Heiden, Switzerland), collected and washed with FACS buffer (2% Fetal Bovine Serum in PBS)(Sigma, St. Louis, MO, USA). The splenocytes were spun down at 300×g for 7 minutes and resuspended at 10 million cells per ml of 90% fetal bovine serum and 10% DMSO (Millipore, Billerica, MA, USA), and aliquoted into cryotubes. The cell aliquots were placed in Mr. Frosty containers (Thermo Fisher Scientific, Waltham, MA, USA) overnight at −80°C and then placed into a ℓN2 freezer in the vapor phase.
2.3 Fluorescent activated cell sorting (FACS)
A complete, step-by-step protocol detailing the following steps, including FACS and B cell co-culture is provided as a supplemental document. Cryopreserved splenocytes were removed from ℓN2 storage, quickly warmed in a 37°C water bath until just thawed, and resuspended in 10 ml prewarmed RPMI with 10% FBS (RPMI complete: RPMI-c). After centrifugation and resuspension in FACS buffer, splenocytes were passed through a negative B cell selection column (StemCell Technologies, Inc., Tukwila, WA, USA), removing CD4+ and CD8+ T cells. This selection resulted in a cell population that was approximately 97% B cells. After B cell selection, cells were washed with FACS buffer and resuspended in 100 μl FACS buffer. These B cells were stained with the decoy stain for 10 minutes at room temperature. The decoy allows identification of B cells that recognize the HIS-tag or streptavidin, so that those non-specific B cells can be discarded during FACS. After incubation with decoy, the target stain and antibodies (B220-PacBlue, CD38-APC, IgM-FITC, and IgD-AF700; BioLegend, San Diego, CA, USA) were added and incubated at 4°C for 25 minutes. Cells were washed with 5 ml of FACS buffer, resuspended at a final concentration of 1.5×107 cells per ml, and passed through a 70-μm filter (Sefar, Heiden, Switzerland) before sorting on a BD FACSAria II with a 100-μm nozzle running at 20 psi. Cells were sorted into 1 ml of IMDM (Thermo Fisher, Waltham, MA, USA) with 10% FBS (IMDM-c) (Gemini Bio-Products, West Sacramento, CA, USA).
2.4 B cell culture and screening
Our culture strategy was to co-culture FACS-sorted antigen positive B cells with CD40L-expressing cells in the presence of IL-4, lipopolysaccharide (LPS), and 2-mercaptoethanol (βME). Irradiated 3T3-msCD40L cells (obtained from the NIH AIDS Reagent Program, contributed by Dr. Mark Connors) (31) were removed from ℓN2 storage, quickly thawed in a 37°C water bath, and washed in 10 ml warm RPMI. After centrifugation, cells were resuspended in IMDM-c and set aside. For each 96-well plate, the outer wells (A1-A12, H1-H12, B1-G1, and B12-G12) were filled with 200 μl sterile dH2O to prevent evaporation from the culture wells, leaving 60 wells in the middle of the plate. 54 wells were used for culturing sorted B cells, and 6 wells contained only 3T3-msCD40L cells, as a control. A 5x stock solution was created with 0.5 ng/ml murine IL-4 (BioLegend, San Diego, CA, USA), 100 μg/ml LPS (Sigma-Aldrich, St. Louis, MO, USA), 250 μM βME (VWR, Radnor PA, USA), and 5×105 irradiated 3T3-msCD40L cells in a final volume of 1.2 ml IMDM-c. 120 μl of this mixture was set aside for use in the control wells containing only feeder cells. The volume of sorted B cells equal to a final well density of 5 or 10 cells was calculated and added to the 5x mixture. The mixture was then diluted to 1x by adding IMDM-c to a final volume that enabled the addition of 100 μl to each culture well (containing an equivalent of either 5 or 10 B cells). This density of B cells typically results in approximately 10–30% ELISA-positive wells. For control wells, the 120 μl of 5x mixture was brought up to 600 μl with IMDM-c, and 100 μl was added to each well. Volumes were scaled as necessary for multiple plates. Plates were placed in a 37°C incubator at 5% CO2 and incubated with minimal disturbance for 12 days.
On days 12 and 14, 10 μl of culture supernatant was removed from each well and tested for antigen binding by capture ELISA. In some cases, we have observed positive ELISA results in as few as 8 days. However, in our experience, 12 days was sufficient to capture secretion in all of our sorting experiments. Capture ELISAs were performed as follows: 50 ng per well of streptavidin (NEB, Ipswich, MA, USA) was plated onto Immulon 2HB plates (Thermo Fisher, San Diego, CA, USA) in 0.1 M sodium bicarbonate (pH 9.4) and incubated at room temperature overnight. The plates were washed with PBS + 0.02% Tween-20 (ELISA wash buffer) and blocked with PBS, 3% BSA for one hour at 37°C. After each incubation step, plates were washed three times in ELISA wash buffer. Biotinylated PfTRAP was added at 200 ng per well in dilution buffer of PBS, 0.2% BSA, and incubated for one hour at 37°C. The plates were then blocked with PBS, 10% nonfat milk, 0.3% Tween-20. After blocking, sample containing either culture supernatant or purified control antibody was added for one hour at 37°C and bound antibodies were detected with either a goat anti-Mouse IgG-HRP polyclonal secondary antibody (Sigma-Aldrich, St. Louis, MO, USA) or a goat anti-Mouse Ig-HRP polyclonal secondary antibody (BD Biosciences, San Jose CA, USA) for one hour at 37°C. After a final wash, 50 μl TMB ELISA substrate (KPL Inc., Gaithersburg, MD, USA) was added and the plates were allowed to develop for three minutes. Development was stopped with equal volume 1M H2SO4 and absorbance at 450 nm was read on a BioTek ELX800 plate reader (BioTek, Winooski, VT, USA).
Following ELISA identification of antigen binding wells, culture supernatant was removed and transferred into new 96-well plates for storage at 4°C. Wells positive for antigen binding were lysed with 100 μL RLT lysis buffer (Qiagen, Hilden, Germany) supplemented with 1 μL 2-mercaptoethanol to prepare the sample for RNA extraction. The lysate was transferred into 1.5 ml Eppendorf tubes and stored at −80°C.
2.5 5′RACE rescue of heavy and light chain sequences
One feature of our protocol is the ability to amplify heavy and light chains from harvested cells by 5′RACE, rather than needing a large set of multiplex primers. To isolate Ig chains, RNA from antigen-binding wells was extracted using the All-Prep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) and approximately 50–250 ng of RNA was used to generate 5′RACE-ready cDNA. 5′RACE-ready cDNA was generated using the SMARTscribe Reverse Transcriptase from Takara-Clontech (Mountain View, CA, USA) according to the manufacturer’s recommended protocol, with the exception of the inclusion of gene-specific primers Mm IgG, Mm IgK, and Mm IgL (see Supplemental Table S1) for the creation of race-ready cDNA. After race-ready cDNA synthesis, cDNAs were diluted in Tris-EDTA and used as a template for PCR. PCR was performed using Q5 polymerase (NEB, Ipswich, MA, USA) in a volume of 25 μl containing 120 nM of SMARTer step-out and Mm IgG CH1 or Mm IgK CH1 reverse primers (depending on heavy or light chain amplifications) and 1.0 μl of race-ready cDNA. PCR cycling conditions were as follows: 98°C – 30s; 30 cycles – 98°C – 10s, 63°C – 30s, 72°C – 30s followed by a 5-minute, final extension at 72°C. PCR amplicons were subsequently visualized and gel-purified using Flash Gels (Lonza, Walkersville, MD, USA), according to the manufacturer’s protocol. Purified PCR amplicons were further adapted by 15 cycles of PCR (same cycling conditions, as above) using step-out primers in order to make them compatible with Gibson assembly into the expression vectors, and then gel-purified again.
2.6 Assembly into mAb IgV-acceptor construct expression vectors
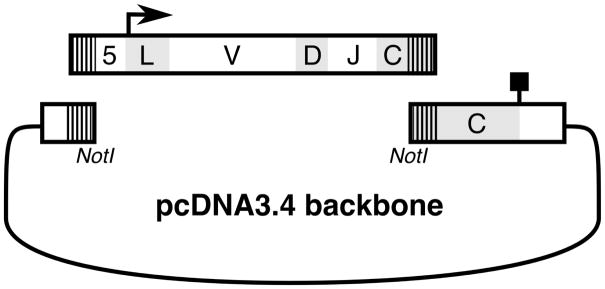
One key advance for this method was the development of an efficient, one step cloning step that enabled both sequencing and protein expression, a process that usually takes multiple days, two cloning steps, and multiple sequencing reactions. This was accomplished by leveraging the universal 5′RACE primer that is present in all amplicons to create a 5′ vector overlap that, together with the overlap in the constant region allows for direct Gibson assembly (Fig. 2). To create the vectors, sequences encoding constant regions of murine IgG (BAQ25543, residues 151–460) and IgK (4IJ3_B, residues 107–214) were codon-optimized for expression in human cells and synthesized as gBlocks (Integrated DNA Technologies, Inc.; Coralville, IA). Additionally, the 5′ end of each construct was designed to contain a NotI restriction site for linearization of the vector to act as an acceptor in a Gibson cloning reaction. This cassette was cloned into a pcDNA3.4 vector (Thermo Fisher, Waltham, MA, USA).
Figure 2. Ligation-independent cloning of antibody genes.
Heavy and light chain PCR amplicons (V-D-J schematic) are amplified containing vector-specific overhangs (hashed lines). Amplicons are then used in a Gibson assembly reaction to create heavy and light chain expression plasmids. 5: 5′UTR, bold arrow: translational start site, L: leader, V: variable region, D: D-region (heavy chain only), J: J-region, C: constant region, bold peg: stop site.
For the Gibson reaction, NEBuilder 2X HiFi DNA Assembly Master Mix (NEB, Ipswich, MA, USA) was used to assemble the purified, adapted PCR amplicons into the appropriate heavy- or light-chain acceptor vector encoding in-frame constant-regions of murine IgG1 or IgK, respectively. After bacterial transformation and selection on antibiotic plates, single clones were screened by Sanger sequencing to assure they contain valid, in-frame immunoglobulin sequences. Sequence analysis of antibody rearrangements was carried out using IgBLAST and IMGT (32, 33). Clones containing a full, in-frame Ig sequence with a valid leader sequence were transfected directly into HEK293 cells for expression of the recombinant antibody. In some cases, a culture well may yield more than one heavy or light chain. In those rare cases, expression of combinations of H + L chains from that well rapidly identifies the correct, functional pairing (see below).
2.7 Expression and purification of recombinant mAbs
Plasmid DNA encoding the heavy and light chains were co-transfected into HEK293 suspension cells using 293-Free Transfection Reagent (EMD-Millipore, Darmstadt, Germany), according to manufacturer’s protocol. After culturing for four days at 37°C, 5% CO2 on a shaker platform, the cells were removed from the supernatant by centrifugation. The mAbs were purified by adjusting the clarified culture supernatant to pH 6.0 with acetic acid and passing it over Protein G Sepharose resin (GE Healthcare, Chicago, IL, USA). Bound antibodies were eluted with 0.1 M glycine, pH 2.7, and then neutralized with 1 M dibasic sodium phosphate. The purified antibodies were buffer exchanged into PBS, pH 7.4, 0.02% sodium azide for storage. Species and purity were verified by analytical size exclusion chromatography (SEC) on a Superdex 200 10/300 column (GE Healthcare, Chicago, IL, USA).
2.8 Antibody binding assays and KD determination
Antibody binding was analyzed by biolayer interferometry (BLI) using an Octet QKe instrument (ForteBio, Inc., Menlo Park, CA, USA). Anti-Mouse IgG Fc Capture (AMC) probes were used to immobilize recombinant antibody directly from the culture supernatant (see above), followed by equilibration in 10x Kinetics Buffer (PBS + 0.1% BSA, 0.02% Tween-20 and 0.05% sodium azide). The mAb-derivatized probes were then dipped into indicated concentrations of antigen in 10X Kinetics Buffer, followed by an incubation step in antigen-free 10X Kinetics Buffer. The resulting sensorgrams were used to generate a global fit using a simple 1:1 binding model using the ForteBio Data Analysis software (version 7.0.1.5), yielding estimated on- and off-rates, as well as the derived KD estimates.
3. Results
3.1 Generation and isolation of PfTRAP-specific B cells
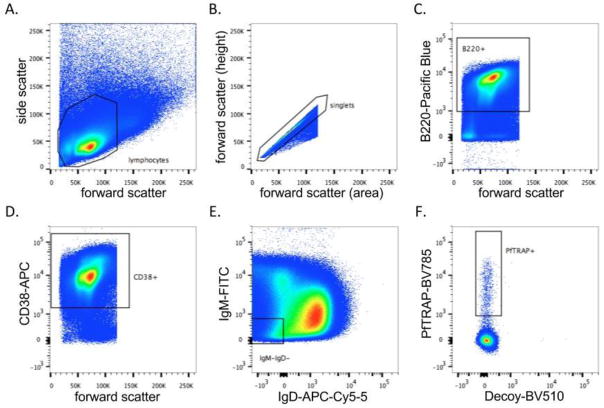
To generate and isolate B cells specific for PfTRAP, we implemented the efficient and cost-friendly workflow that is summarized in Fig. 1. Balb/c mice were immunized as outlined above to generate antigen-specific responses, and the final boost was performed two days before spleen harvest and sorting. Spleens were harvested and processed on the same day for B cell FACS. 288 million splenocytes were passed over a B cell isolation column to remove any T cells (CD3+ cells), resulting in 71 million B cells. These cells were stained with cell surface marker antibodies and antigens (Table 1), and sorted for the PfTRAP+, Decoy (PyCSP)−, IgM−, IgD−, B220+, CD38+ population (Fig. 3), a population consisting of antigen-educated, class- switched B cells. Sorting yielded 5017 PfTRAP+ B cells, 2430 of which were plated over the irradiated 3T3-msCD40L fibroblast cells at a density of either 5 or 10 cells per well.
Table 1. FACS reagents.
List of the antibodies and fluorophore conjugates used in this protocol, including the dye, commercial source, and catalog number. When available, the antibody registry number is listed.
| MARKER | DYE | SOURCE | CATALOG NO. | ANTIBODY REGISTRY |
|---|---|---|---|---|
| anti-PfTRAP | BV785 | BioLegend | 405249 (for BV785-strep) | N/A |
| IgD | AF700 | BioLegend | 405730 | AB_2563341 |
| CD38 | APC | BioLegend | 102712 | AB_312933 |
| IgM | FITC | BioLegend | 406506 | AB_315056 |
| Decoy (anti-PyCSP) | BV510 | BioLegend | 405233 (for BV510-strep) | N/A |
| B220/CD45R | PacBlue | BioLegend | 103227 | AB_492876 |
Figure 3. Isolation of PfTRAP antigen-positive B cells.
Splenocytes were harvested from PfTRAP-immunized mice, subjected to B cell enrichment, and stained for FACS analysis. Gates were set as follows: A) total lymphocytes, B) cell singlets, C) B220+ B cells (Pacific Blue), D) CD38+ (APC), E) IgM– and IgD– (FITC and AF700, respectively), and F) PfTRAP+ (BV785) and irrelevant protein decoy– (BV510).
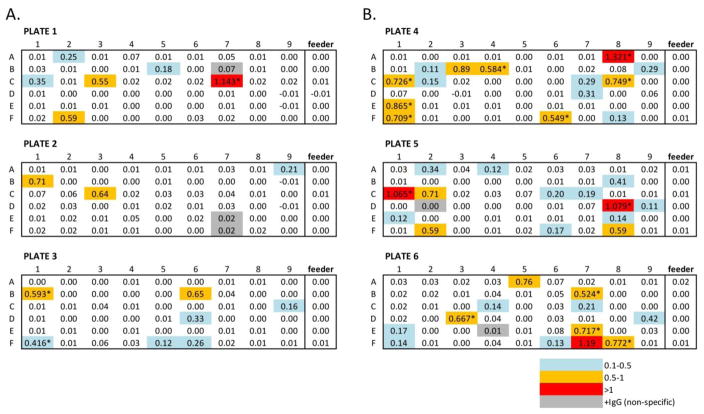
Plated cultures were monitored for the secretion of anti-PfTRAP antibodies by capture ELISA on days 12 and 14. Approximately 10% of the wells seeded with 5 cells per well showed significant binding to PfTRAP by capture ELISA, whereas 24.7% were similarly scored from the 10-cells/well group (Fig. 4). Soluble immunoglobulin (Ig) production was used as a proxy for cell survival/proliferation in B cell culture. In this experiment, we identified a total of five wells that expressed IgG, but did not bind to PfTRAP (i.e., 90% of IgG+ wells bound to target antigen). These results illustrate that this protocol can efficiently identify, harvest and culture a significant number of B-cells capable of antigen recognition from spleens of immunized mice.
Figure 4. Capture ELISA analysis of PfTRAP antigen binding.
Capture ELISA was used to assess the binding capacity of B cell culture supernatants. A) B cells were plated at 5 cells/well. B) B cells were plated at 10 cells/well. Cell color indicates antigen binding, measured as the OD at 450 nm: blue (0.1–0.5), yellow (0.5–1.0), and red (>1.0). Grey shaded wells indicate that IgG expression was detected in the absence of target binding. Asterisks indicate wells that were tested and removed for storage at day 12, the remainder of the wells were tested at day 14.
3.2 Antibody rescue, expression, and binding characteristics
Six PfTRAP ELISA-positive wells were selected for mAb rescue, designated AKBR1-6. Six microliters of purified RNA were used for the production of RACE-ready cDNA, which was followed by PCR (primers in Supplementary Table S1). The initial PCR reaction yielded an amplicon of approximately 550 base pairs (bp) for the heavy chain, including the entire V-D-J rearrangement and a portion of the constant region. The kappa light chain amplicon was approximately 500 bp and included the entire V-J rearrangement and a portion of the kappa constant region (Supplementary Fig. S1).
After Gibson assembly, expression plasmids containing anti-PfTRAP heavy and light chains were transformed into E. coli and screened for inclusion of heavy and light chains. Plasmids containing inserts of the expected size were validated by Sanger sequencing, identifying a variety of heavy- and light-chain gene usage (Tables 2,3). Heavy chains possessed CDRH3 lengths ranging from 9–14 amino acids. The light chains were less variable, with CDRL3s of 8–9 amino acids. Both CDR3 length ranges are consistent with antibodies generated in other model systems (34, 35). Each of the six heavy and light chain pairs were co-transfected into HEK293 cells to express recombinant antibodies. Protein G purifications of small-scale transfections yielded a range of recombinant antibody: from 20–200 μg from a 10-mL culture (Supplementary Table S2).
Table 2. Heavy chain gene annotation.
Heavy chain sequences were annotated for V, D, and J gene usage and for CDRH3 length. The most probable match to known gene segments is listed in the table. When two or more genes are equally probable, all matches are listed. The CDRH3 length is the number of amino acids in the CDRH3 as identified by IMGT and IgBLAST.
| AKBR | WELL | V | D | J | CDRH3 |
|---|---|---|---|---|---|
| 1 | 1C7 | IGHV1S29*02 | IGHD6-1*01,IGHD6-2*01,IGHD6-2*02 | IGHJ3*01 | 9 |
| 2 | 3B1 | IGHV8-12*01 | IGHD2-3*01 | IGHJ4*01 | 13 |
| 3 | 3F1 | IGHV3-2*02 | IGHD1-1*01 | IGHJ2*01 | 15 |
| 4 | 5C1 | IGHV1-69*01, IGHV1-69*02 | IGHD2-3*01 | IGHJ4*01 | 14 |
| 5 | 5D8 | IGHV6-6*01 | IGHD3-3*01,IGHD4-1*01,IGHD4-1*02 | IGHJ4*01 | 11 |
| 6 | 4A8 | IGHV8-12*01 | IGHD2-3*01 | IGHJ1*01 | 15 |
Table 3. Light chain gene annotation.
Light chain sequences were annotated for V and J gene usage and for CDRL3 length. The most probable match to known gene segments is listed in the table. The CDRH3 length is the number of amino acids in the CDRH3 as identified by IMGT and IgBLAST.
| AKBR | WELL | V | J | CDRL3 |
|---|---|---|---|---|
| 1 | 1C7 | IGKV3-4*01 | IGKJ2*01 | 9 |
| 2 | 3B1 | IGKV12-41*01 | IGKJ2*01 | 8 |
| 3 | 3F1 | IGKV4-59*01 | IGKJ5*01 | 9 |
| 4 | 5C1 | IGKV17-127*01 | IGKJ5*01 | 9 |
| 5 | 5D8 | IGKV1-110*01 | IGHJ4*01 | 8 |
| 6 | 4A8 | IGKV12-46*01 | IGKJ1*01 | 9 |
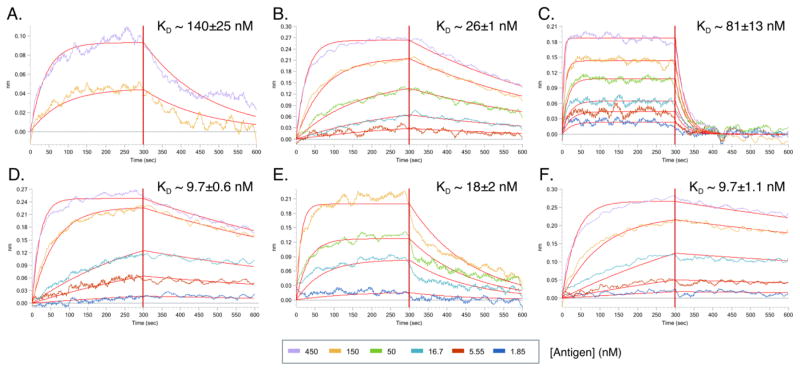
To assess the functionality of the cloned anti-PfTRAP mAbs, the binding capability and dissociation constants (KD) of each recombinant mAb was assessed by BLI (Fig. 5, Supplementary Table S3). The mAbs AKBR1–6 exhibited a range of KD values, varying from 9.73×10−9 to 1.40×10−7 M. Thus, we were able to use this method to retrieve and produce six unique, recombinant mAbs (out of six selected culture wells) that utilize different V, D and J gene segments, and are able to recognize the cognate recombinant antigen with a spectrum of affinities.
Figure 5. BLI analysis of recombinant anti-PfTRAP mAbs.
AMC biosensors derivatized with recombinant mAbs were incubated for 300 s in well solutions containing serially diluted PfTRAP to produce color-coded sensorgrams (colors correspond to the PfTRAP concentrations, see legend). The sensors were then transferred into solutions without PfTRAP and the complexes were allowed to dissociate for another 300 s. Each of the datasets was globally fitted to produce the red trendlines and the estimated on- and off-rates from which KD estimates were calculated. A) AKBR1, B) AKBR2, C) AKBR3, D) AKBR5, E) AKBR4, F) AKBR6.
4. Discussion
In this report, we describe a detailed protocol for efficient generation of mAbs from immunized mice. The key features of this protocol are 1) recovery of antigen-binding B cells using FACS, 2) culture of primary B cells for the purpose of proliferation and antibody secretion, 3) secondary screening of antibody-containing B cell culture supernatants, and 4) optimized 5′RACE-based recovery of heavy- and light-chain sequences into an expression cassette containing constant-region sequences. This protocol can be completed in ~20 days after the immunization protocol ends, and can result in identification of multiple unique mAbs for further characterization, as we have shown here.
As noted previously, selection for antigen-binding B cells by FACS as a starting point for mAb generation is a major advance over the traditional random fusion of murine splenocytes, and has become a mainstay of single B cell cloning. Here we also suggest adding a pre-FACS negative selection of B cells to remove T cells, as we found B cells to make up approximately 25% of total splenocytes. In our hands, this step helped sorting efficiency and improved mAb recovery. Furthermore, as previously described (36), decoy staining within our gating strategy (Fig. 3) can be used to eliminate nonspecific binding to the target Ag, especially those B cells that may recognize the HIS-tag on the target antigen or have nonspecific binding to streptavidin. As illustrated in Fig. 3, only ~0.007 % of the total B cells (or 0.63% of B220+, IgM/D-, CD38+ B cells) recovered from the spleen were capable of recognizing the target Ag. Thus, an optimized FACS protocol is a key parameter to ensure successful mAb isolation.
We utilized B cell culture in this protocol to ensure the efficient identification of antigen-specific B cell wells for mAb rescue. A strength of this approach is that it allows the identification of antigen specific B cells wells before PCR and protein production steps become necessary. Additionally, at this step different screening antigens can be used to bin for specific epitopes. However, we found that murine primary B cell culture presented significant challenges and necessitated development of new culture conditions, as differences in growth requirements between murine and human B cells prevented us from adapting well-established human culture conditions (31). While our culture conditions permit us to recover a significant number of Ag-positive wells, a large percentage of the wells did not give rise to expanded immunoglobulin-producing B cells (Fig. 4). This suggests that further optimization of culture conditions may achieve better B cell survival and increase the efficiency of Ag-binding B cell recovery.
Another crucial aspect is the approach used to screen for antigen positive B cell culture wells. The secondary screen in our protocol employs streptavidin-capture ELISA to immobilize the biotinylated target Ag on the plate, instead of the more traditional direct immobilization on pretreated plastic. This mode of ELISA previously allowed us to identify mAbs that recognize conformationally sensitive epitopes that were undetectable with directly-immobilized target Ag (data not shown). Among the mAbs in this report, recognition of PfTRAP by all of the mAbs but one (AKBR5) was significantly impaired or ablated by the direct immobilization PfTRAP, indicating that their epitopes are either perturbed or destroyed by adsorption to the plate (Supplementary Fig. S2). This observation indicates that direct-immobilization ELISA may not faithfully identify B cell wells that contain antibodies targeting some conformational epitopes, and that streptavidin-capture or a similar target-orienting technique is critical in order to identify the maximal number of B cell specificities.
A key advance for this protocol was the development of an efficient 5′RACE-based recovery strategy that allowed us to create finalized expression constructs for heavy and light chains in one cloning step using Gibson assembly. Typically this has been done as a two-step process consisting of the initial PCR cloning step followed by sequence dependent sub-cloning, as diversity within the Ig leader segments and V genes necessitates sequencing the rescued immunoglobulin before it can be subcloned into an expression vector. By our method, the PCR product can be assembled straight into the expression vector, and importantly, the method is independent of leader sequence or V gene usage. In our experience, using this strategy can cut an average of 3–4 days off of similar protocols using two step cloning. In our hands, this procedure has a high success rate, allowing us to successfully recover 6 non-clonal functional mAbs from 6 different ELISA-positive wells in this study. However, two considerations are relevant. First, the product usually contains the 5′UTR and the native leader sequence, the composition of which may impact the expression efficiency of the mAb. Second, incomplete first strand synthesis can result in a sequence that lacks a leader sequence and start codon, resulting in a non-functional sequence. Thus, there are rare cases in which additional repair cloning steps may be required prior to attempting expression of recombinant mAbs. However, in the rare cases in which we observed these phenomena, simply screening more clones resulted in recovery of full length, functional Ig sequences.
The variety of recovered sequences (Tables 2 and 3) show a diverse pattern of gene usage in heavy- and light-chain sequences with unique CDR3 sequences. This suggests that the B cell receptor recovery via 5′RACE was unbiased, and that this method should be able to rescue mAbs from a variety of V genes. Furthermore, the observed genetic diversity, together with different sensitivities to ELISA conditions (see capture-ELISA discussion above) and diverse Ag-binding characteristics (Fig 5), all suggest that the recovered mAbs described here likely recognize different epitopes on PfTRAP. Specific epitope mapping will need to be carried out to further support this hypothesis. However, it is clear that this method is well suited to sampling diverse B cell responses against complex protein antigens.
Antibody discovery remains an important tool to dissect B cell mediated immunity, and the methods to do so are continually improving. Here we have described a method for mAb isolation and cloning based on pre-screening mAbs for functionality through B cell co-culture and an efficient PCR rescue and antibody production scheme. Although different antigens presented different degrees of challenge with respect to mAb isolation and cloning, we have successfully deployed this workflow in our ongoing efforts to identify protective mAbs directed against the malaria parasite. To date we have used this method to isolate a number of mAbs against eleven different recombinant antigens derived from three different species of Plasmodium (P. falciparum, P. vivax, P. yoelii - data not shown). However, the power of this method is that it is not dependent on the origin of the antigen, and is a tool that could be easily deployed in the study of any vaccine or infectious disease, or in the development of novel therapeutics in murine model systems.
Supplementary Material
Highlights.
mAb discovery method using FACS identification of antigen-specific B cells.
B cell culture, screening, and rescue by 5′RACE ensures efficiency and accuracy.
Co-culture of murine B cells to ensure efficient well selection with functional prescreening.
Efficient mAb rescue allows rapid analysis of mAb binding and affinity.
Method produces diverse mAbs with a range of specificities and binding affinities.
Acknowledgments
We gratefully acknowledge the Vivarium staff at the Center for Infectious Disease Research (CIDR), including Tim Dawe. We also thank Nick Lejarcegui and Weldon DeBusk at the Flow Cytometry Core at CIDR for assistance with cell sorting. Additional thanks to M. Pepper and A. Krishnamurty for initial technical assistance with FACS. This study was funded through a CIDR internal grant to DNS and NIH/NIAID grant R01AI117234 to SHK and DNS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Corti D, Lanzavecchia A. Efficient Methods To Isolate Human Monoclonal Antibodies from Memory B Cells and Plasma Cells. Microbiol Spectr. 2014:2. doi: 10.1128/microbiolspec.AID-0018-2014. [DOI] [PubMed] [Google Scholar]
- 2.Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 3.Mann CJ. Rapid isolation of antigen-specific clones from hybridoma fusions. Nat Methods. 2007:4. [Google Scholar]
- 4.Coller HA, Coller BS. Poisson statistical analysis of repetitive subcloning by the limiting dilution technique as a way of assessing hybridoma monoclonality. Methods Enzymol. 1986;121:412–417. doi: 10.1016/0076-6879(86)21039-3. [DOI] [PubMed] [Google Scholar]
- 5.Yokoyama WM, Christensen M, Dos Santos G, Miller D, Ho J, Wu T, Dziegelewski M, Neethling FA. Production of monoclonal antibodies. Curr Protoc Immunol. 2013;102(Unit 2.5) doi: 10.1002/0471142735.im0205s102. [DOI] [PubMed] [Google Scholar]
- 6.Smith SA, Crowe JE., Jr Use of Human Hybridoma Technology To Isolate Human Monoclonal Antibodies. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.AID-0027-2014. AID–0027–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoogenboom HR. Overview of antibody phage-display technology and its applications. Methods Mol Biol. 2002;178:1–37. doi: 10.1385/1-59259-240-6:001. [DOI] [PubMed] [Google Scholar]
- 8.Ogunniyi AO, Thomas BA, Politano TJ, Varadarajan N, Landais E, Poignard P, Walker BD, Kwon DS, Love JC. Profiling human antibody responses by integrated single-cell analysis. Vaccine. 2014;32:2866–2873. doi: 10.1016/j.vaccine.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith K, Garman L, Wrammert J, Zheng N-Y, Capra JD, Ahmed R, Wilson PC. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc. 2009;4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng N-Y, Mays I, Garman L, Helms C, James J, Air GM, Capra JD, Ahmed R, Wilson PC. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 12.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiller T, Busse CE, Wardemann H. Cloning and expression of murine Ig genes from single B cells. J Immunol Methods. 2009;350:183–193. doi: 10.1016/j.jim.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Volkheimer AD, Weinberg JB, Beasley BE, Whitesides JF, Gockerman JP, Moore JO, Kelsoe G, Goodman BK, Levesque MC. Progressive immunoglobulin gene mutations in chronic lymphocytic leukemia: evidence for antigen-driven intraclonal diversification. Blood. 2007;109:1559–1567. doi: 10.1182/blood-2006-05-020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR Protocol G. Principal Investigators. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P Protocol G. Principal Investigators. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao H-X, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang K-K, Yang Y, Chen X, Gao F, Munshaw S, Kepler TB, Denny T, Moody MA, Haynes BF. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koelsch K, Zheng N-Y, Zhang Q, Duty A, Helms C, Mathias MD, Jared M, Smith K, Capra JD, Wilson PC. Mature B cells class switched to IgD are autoreactive in healthy individuals. J Clin Invest. 2007;117:1558–1565. doi: 10.1172/JCI27628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng W, Li L, Xiong W, Fan X, Deng H, Bett AJ, Chen Z, Tang A, Cox KS, Joyce JG, Freed DC, Thoryk E, Fu T-M, Casimiro DR, Zhang N, Vora AK, An Z. Efficient generation of monoclonal antibodies from single rhesus macaque antibody secreting cells. MAbs. 2015;7:707–718. doi: 10.1080/19420862.2015.1051440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kivi G, Teesalu K, Parik J, Kontkar E, Ustav M, Jr, Noodla L, Ustav M, Männik A. HybriFree: a robust and rapid method for the development of monoclonal antibodies from different host species. BMC Biotechnol. 2016;16:2. doi: 10.1186/s12896-016-0232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Boehmer L, Liu C, Ackerman S, Gitlin AD, Wang Q, Gazumyan A, Nussenzweig MC. Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat Protoc. 2016;11:1908–1923. doi: 10.1038/nprot.2016.102. [DOI] [PubMed] [Google Scholar]
- 22.Corsiero E, Bombardieri M, Carlotti E, Pratesi F, Robinson W, Migliorini P, Pitzalis C. Single cell cloning and recombinant monoclonal antibodies generation from RA synovial B cells reveal frequent targeting of citrullinated histones of NETs. Ann Rheum Dis. 2016;75:1866–1875. doi: 10.1136/annrheumdis-2015-208356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaushansky A, Douglass AN, Arang N, Vigdorovich V, Dambrauskas N, Kain HS, Austin LS, Sather DN, Kappe SH. Malaria parasites target the hepatocyte receptor EphA2 for successful host infection. Science. 2015;350:1089–1092. doi: 10.1126/science.aad3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harupa A, Sack B, Lakshmanan V, Arang N, Douglass AN, Oliver BG, Stuart AB, Sather N, Lindner SE, Hybiske K, Torii M, Kappe SH. SSP3 is a novel Plasmodium sporozoite surface protein with a role in gliding motility. Infect Immun. 2014 doi: 10.1128/IAI.01800-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang JY, Song WT, Li Y, Chen WJ, Yang D, Zhong GC, Zhou HZ, Ren CY, Yu HT, Ling H. Improved expression of secretory and trimeric proteins in mammalian cells via the introduction of a new trimer motif and a mutant of the tPA signal sequence. Appl Microbiol Biotechnol. 2011;91:731–740. doi: 10.1007/s00253-011-3297-0. [DOI] [PubMed] [Google Scholar]
- 26.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carbonetti S, Oliver BG, Glenn J, Stamatatos L, Sather DN. Soluble HIV-1 envelope immunogens derived from an elite neutralizer elicit cross-reactive V1V2 antibodies and low potency neutralizing antibodies. PLoS One. 2014;9:e86905. doi: 10.1371/journal.pone.0086905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Backliwal G, Hildinger M, Hasija V, Wurm FM. High-density transfection with HEK-293 cells allows doubling of transient titers and removes need for a priori DNA complex formation with PEI. Biotechnol Bioeng. 2008;99:721–727. doi: 10.1002/bit.21596. [DOI] [PubMed] [Google Scholar]
- 29.Keitany GJ, Sack B, Smithers H, Chen L, Jang IK, Sebastian L, Gupta M, Sather DN, Vignali M, Vaughan AM, Kappe SH, Wang R. Immunization of mice with live-attenuated late liver stage-arresting Plasmodium yoelii parasites generates protective antibody responses to preerythrocytic stages of malaria. Infect Immun. 2014;82:5143–5153. doi: 10.1128/IAI.02320-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keitany GJ, Kim KS, Krishnamurty AT, Hondowicz BD, Hahn WO, Dambrauskas N, Sather DN, Vaughan AM, Kappe SHI, Pepper M. Blood Stage Malaria Disrupts Humoral Immunity to the Pre-erythrocytic Stage Circumsporozoite Protein. Cell Rep. 2016;17:3193–3205. doi: 10.1016/j.celrep.2016.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J, Doria-Rose NA, Longo NS, Laub L, Lin CL, Turk E, Kang BH, Migueles SA, Bailer RT, Mascola JR, Connors M. Isolation of human monoclonal antibodies from peripheral blood B cells. Nat Protoc. 2013;8:1907–1915. doi: 10.1038/nprot.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brochet X, Lefranc M-P, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–8. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steiniger SCJ, Dunkle WE, Bammert GF, Wilson TL, Krishnan A, Dunham SA, Ippolito GC, Bainbridge G. Fundamental characteristics of the expressed immunoglobulin VH and VL repertoire in different canine breeds in comparison with those of humans and mice. Mol Immunol. 2014;59:71–78. doi: 10.1016/j.molimm.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 35.Vigdorovich V, Oliver BG, Carbonetti S, Dambrauskas N, Lange MD, Yacoob C, Leahy W, Callahan J, Stamatatos L, Sather DN. Repertoire comparison of the B-cell receptor-encoding loci in humans and rhesus macaques by next-generation sequencing. Clin Transl Immunology. 2016;5:e93. doi: 10.1038/cti.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor JJ, Martinez RJ, Titcombe PJ, Barsness LO, Thomas SR, Zhang N, Katzman SD, Jenkins MK, Mueller DL. Deletion and anergy of polyclonal B cells specific for ubiquitous membrane-bound self-antigen. J Exp Med. 2012;209:2065–2077. doi: 10.1084/jem.20112272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.