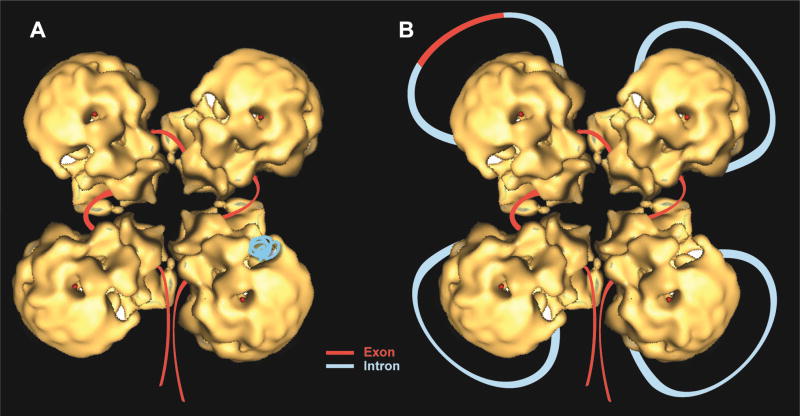
Fig. 13. A refined schematic model of the supraspliceosome.
Correctly orientated native spliceosome volume [37] was placed on a class average, and an idealized supraspliceosome particle was formed by rotating the native volume by 90°, 180° and 270° to generate a four-fold pattern. The rotated volumes fitted well into the parts of neighboring sub-complexes seen in the average image [80]. Schematic models of the supraspliceosome in which the pre-mRNA (introns in blue, exons in red) is connecting four native spliceosomes [29, 37, 80]. The supraspliceosome presents a platform onto which the exons can be aligned and splice junctions can be checked before splicing occurs. (A) The pre-mRNA that is not being processed is folded and protected within the cavities of the native spliceosome. (B) When a staining protocol that allows visualization of nucleic acids was used, RNA strands and loops were seen emanating from the supraspliceosomes [30]. Under these conditions, the RNA kept in the cavity likely unfolded and looped-out. In the looped-out scheme an alternative exon is depicted in the upper left corner.