Figure 1.
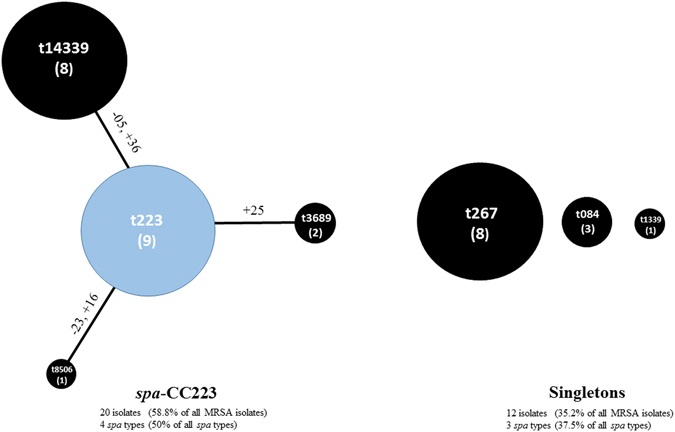
Population structure of the tested MRSA isolates (n = 34) based on BURP analysis. This analysis was performed using the Based Upon Repeat Pattern (BURP) algorithm of the Ridom StaphType software (Ridom GmbH, Würzburg, Germany) at a cost setting of ≤ 5 and excluding spa-types with 5 or fewer repeats. Each dot represents a different spa type, with the diameter of the dot being proportional to the quantity of the corresponding spa type. Clusters of linked spa types correspond to spa clonal complexes (spa-CCs). The predict founder of a cluster (which was used for defining the cluster) is shown in blue, while the others in black. Near the lines of connection, the mutations involved in the transition from a spa type to the next one are reported in detail. All DNA changes are meant to occur from the founder to the periphery. Legend: numbers along the lines refer to the repeat sequence involved in the mutation; +indicates the acquisition of a repeat sequence; - indicates the loss of a repeat sequence; within circles the numbers of the strains of each CC appear between brackets. In summary, the analysis identified a single clonal complex (spa-CC223) comprising spa types t223, t14339, t3689, and t8506; n = 20 isolates, and accounted for 58.8% of all tested MRSA isolates, as well as 3 singletons (t267, t084, and t1339; n = 12 isolates, 35.2%), while excluded 2 isolates (t380, 5.8%) from the clustering, as they consisted of four repeat units only.
