Fig. 4.
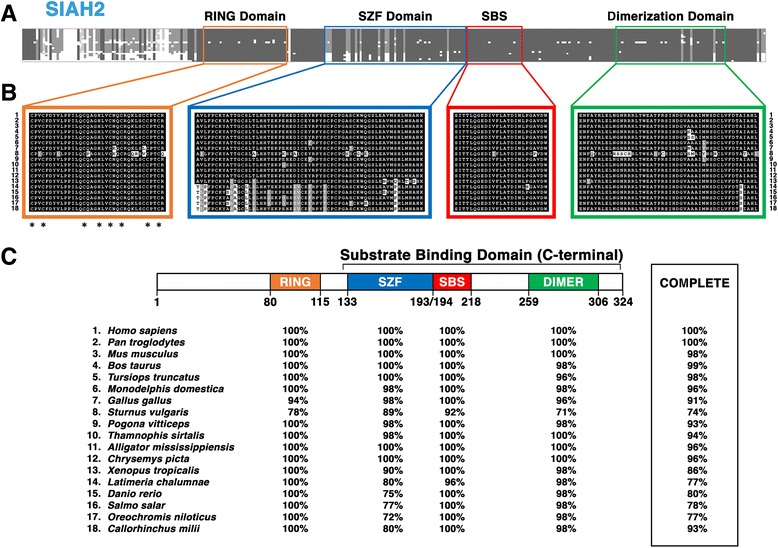
Sequence alignment of the vertebrate SIAH2 subfamily reveals its invariant amino acid residues, and the four conserved structural motifs. Sequence comparison of SIAH2 proteins from 18 representative vertebrate species (#1-#18) is shown. a The level of amino acid divergence in the N-terminal fragments (#1 to #80) is high. Four key functional domains are marked in four distinct colors: RING domain (orange), SZF domain (blue), SBS (red), and DIMER domain (green). b Schematic illustration of amino acid conservation within the 4 domains of the SIAH2 sequences is shown. Amino acid identity is shown as white letters in a black box, amino acid similarity is shown as white letters in a grey box, and amino acid divergence is shown as black letters in a white box. The asterisks located below the RING domain alignment indicate unanimous conservation of the cysteine (Cys)/histidine (His) zinc-binding residues. c The percentages of amino acid conservation in each distinct domain and the entire SIAH2 sequence between human and each of the representative vertebrate species are shown. The diagram of the domain architecture was based on Homo sapiens SIAH2. The SIAH2 sequence for C. milii was incomplete, and these gaps induced by the incompleteness of the sequence were disregarded when calculating conservation across the whole protein
