Fig. 6.
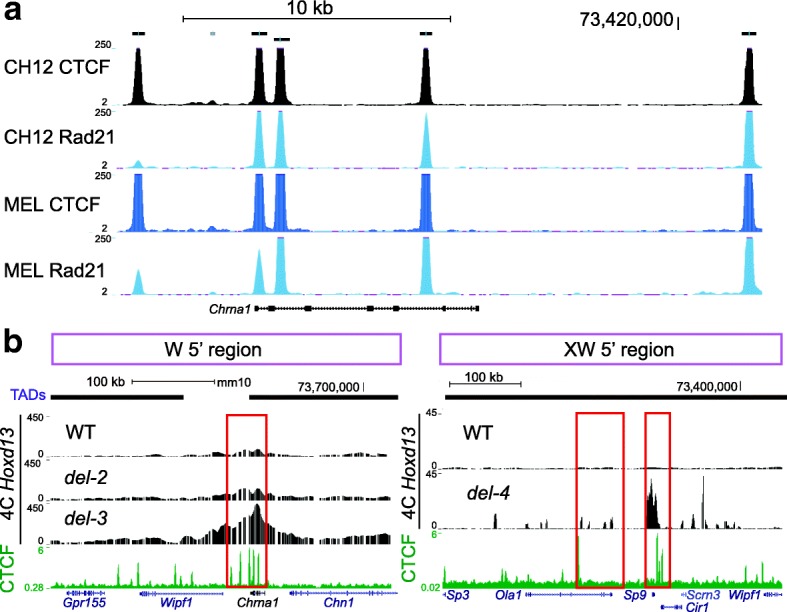
De novo TAD boundaries are CTCF-rich regions. The tracks corresponding to the CTCF and cohesion Chip-seq data were extracted either from ENCODE (a) or from our experiments using distal limb bud cells at E12.5 (b). They are displayed in the UCSC Genome Browser (https://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr2%3A73563281-73580338&hgsid=601487871_7lJaNpYWWLpaaX1lmfxZSqC4FLKC). The regions emphasized are the DNA segments where contacts drastically decrease, as observed in Fig. 5b. WT wild type
