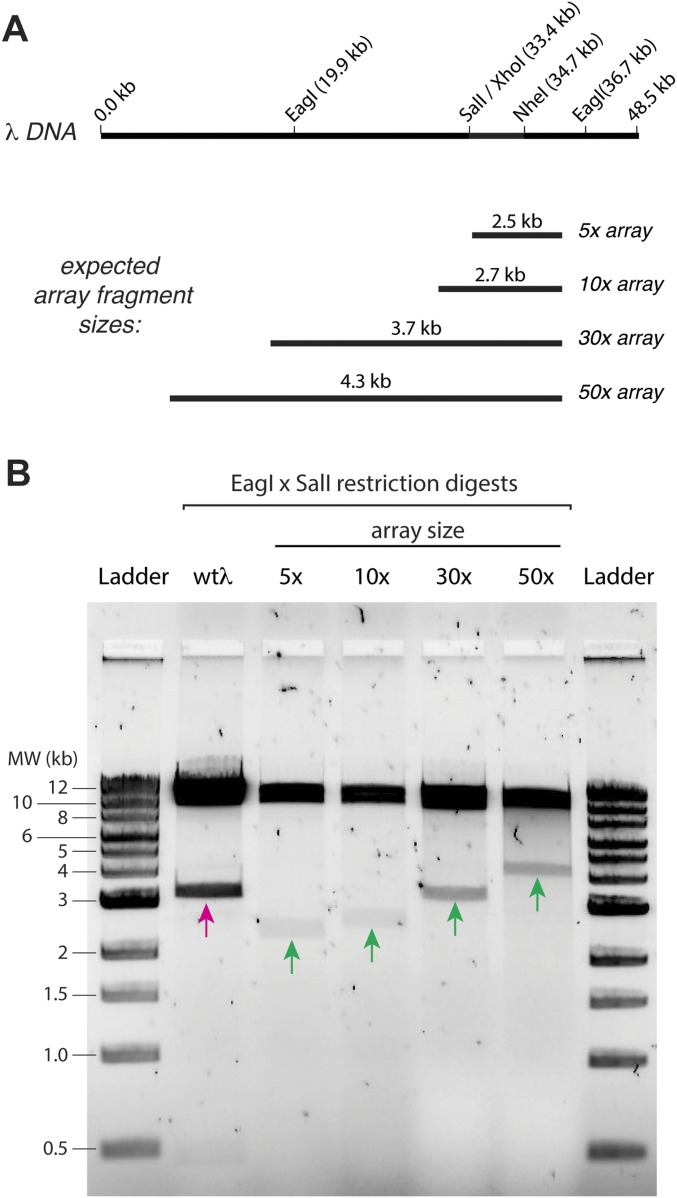
Fig. S3.
Engineered 5×, 10×, 30×, and 50× arrays of EcoRI binding sites on λ-DNA. (A) A schematic of the λ-DNA and a schematic of the expected fragment sizes for the different DNA array lengths when digested with SalI and EagI. (B) A 1% agarose gel showing that the length of engineered sites is as expected. In this assay, 5-μL aliquots of the engineered λ-DNA were incubated with 1 μL of SalI-HF (R3138S; New England BioLabs) and EagI-HF (R3505S; New England BioLabs) in 38 μL water supplemented with 5 μL of 5× CutSmart buffer (B7204S; New England BioLabs) for 2 h. The reaction was mixed with 10 μL of 6× loading dye (B7024S; New England BioLabs) and run on the gel for 1 h at 100 V; a 1-kb DNA ladder (N3232S; New England BioLabs) was also run. The magenta arrow highlights the 3.3-kb native DNA fragment [SalI (33.4 kb) to EagI (36.7 kb)] present before cloning in the array fragments. The green arrows highlight the array-containing fragments for each of the different substrates. MW, molecular weight.