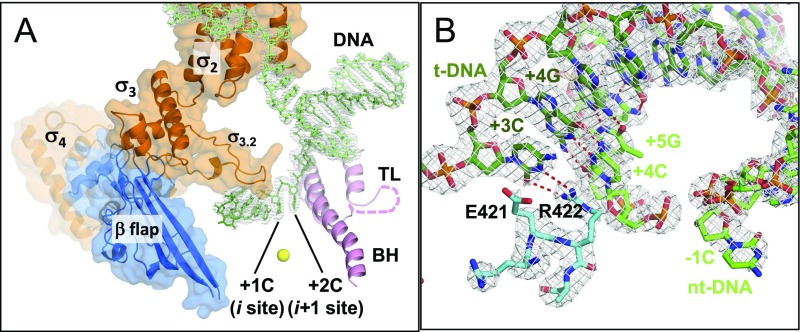
Fig. S1.
Structures of the T. thermophilus RNAP holoenzyme DNA complex mimicking the pyrG promoter open complex. (A) Ribbon models of RNAP domains and motifs are shown with transparent surfaces of the σ-factor and the β-flap domain (TL, trigger loop; BH, bridge helix). DNA strands are shown as stick models (template DNA, dark green; nontemplate DNA, light green). The disordered region of the TL is shown as a dashed line. The nucleotide binding sites (i and i+1) are indicated, and the active site catalytic Mg2+ is shown as a yellow sphere. The gray mesh shows the 2Fo–Fc electron density for the DNA strands contoured at 1.5 σ. (B) Stick models of the downstream DNA and the β-subunit fork loop 2 are shown with their 2Fo–Fc electron densities (gray mesh) contoured at 1.5 σ. Hydrogen bonds between DNA bases and the salt bridge between +3G and R422 are depicted as red dashed lines.